Identification of corticosteroids as potential inhibitor against glycolytic enzyme hexokinase II role in cancer glycolysis pathway: a molecular docking study
Verma Abhishek Kumar, Srivastava Vijay Kumar, Srivastava Sandeep Kumar
Research Articles | Published: 03 February, 2023
Online ISSN : 2229-4473.
Website:www.vegetosindia.org
Pub Email: contact@vegetosindia.org
First Page: 173
Last Page: 180
Views: 610
Keywords:
Corticosteroid drugs, Cancer, Metabolic enzymes, Molecular docking, Protein–ligand interaction
Abstract
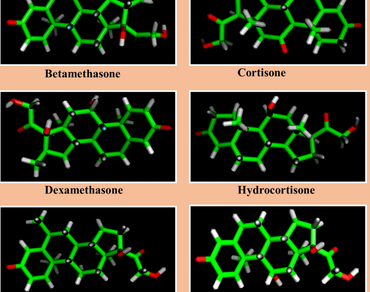
Malignant cells have a significant up-regulation of enzymes that control their bioenergetics and biosynthetic machinery, which is emerging as a cancer hallmark. As a result, new anticancer therapies have centered on targeting metabolic enzymes, resulting in the identification of specific metabolic inhibitors. Corticosteroid medicines (BMS, CS, DMS, HCS, MPS, and PS) are one of these inhibitors, having a broad range of anticancer action due to their ability to inhibit cancer. The molecular characterization of its binding to a wide range of target enzymes, on the other hand, remains mainly elusive. As a result, in the current study, we used molecular modeling, docking and interaction studies to investigate the molecular nature of corticosteroid compounds with important key target enzyme hexokinase II of glycolysis. A comparative analysis of the docking scores with respect to the corticosteroid drugs strongly indicated that both derivatives display efficient binding strength to this target. Furthermore, ADME/T analyses of the drug-likeness of six corticosteroid compounds revealed that all these medicines met desirable drug-like characteristics. The findings of this study shed light on the molecular properties of six corticosteroid medications binding to hexokinase II, which could help in the development and optimization of cancer therapy regimens involving these drugs.
(*Only SPR Members can get full access. Click Here to Apply and get access)
References
Aft RL, Zhang FW, Gius D (2002) Evaluation of 2-deoxy-D-glucose as a chemotherapeutic agent: mechanism of cell death. Br J Cancer 87(7):805–812
Alfarouk KO, Verduzco D, Rauch C, Muddathir AK, Adil HB, Elhassan GO et al (2014) Glycolysis, tumor metabolism, cancer growth and dissemination. A new pH-based etiopathogenic perspective and therapeutic approach to an old cancer question. Oncoscience. 1(12):777
Azam F, Amer AM, Abulifa AR, Elzwawi MM (2014) Ginger components as new leads for the design and development of novel multi-targeted anti-Alzheimer’s drugs: a computational investigation. Drug Des Dev Ther 8:2045
Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, Tang Y (2012) AdmetSAR: a comprehensive source and free tool for assessment of chemical ADMSET properties. Bioinformatics. https://doi.org/10.1021/ci300367a
Ciavardelli D, Rossi C, Barcaroli D, Volpe S, Consalvo A, Zucchelli M et al (2014) Breast cancer stem cells rely on fermentative glycolysis and are sensitive to 2-deoxyglucose treatment. Cell Death Dis 5(7):e1336
Dang CV, Hamaker M, Sun P, Le A, Gao P (2011) Therapeutic targeting of cancer cell metabolism. J Mol Med Berl Ger 89:205–212
DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB (2008) The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7:11–20. https://doi.org/10.1016/j.cmet.2007.10.002
DeLano WL (2002) The PyMOL molecular graphics system. http://www.pymol.org
Discovery Studio Visualization (2016) [Internet]. [Cited 2 Dec 2016]. http://accelrys.com/products/collaborativescience/biovia-discovery-studio/visualization-download.php
Freshney RI, Frame MC, Vaughan PFT, Graham DI (1986) Phenotypic modification of human glioma. Biology of brain tumour. Springer, Boston, pp 35–41
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674. https://doi.org/10.1016/j.cell.2011.02.013
Housman G, Byler S, Heerboth S, Lapinska K, Longacre M, Snyder N et al (2014) Drug resistance in cancer: an overview. Cancers 6:1769–1792. https://doi.org/10.3390/cancers6031769
Jha V, Matharoo DK, Kasbe S, Gharat K, Rathod M, Sonawane N, Kanade T (2021) Computational screening of phytochemicals to discover potent inhibitors against chinkungunya virus. Vegetos 34(3):515–527. https://doi.org/10.1007/s42535-021-00227-9
Khan A, Mohammad T, Shamsi A, Hussain A, Alajmi MF, Husain SA, Hassan MI (2021) Identification of plant-based hexokinase 2 inhibitors: combined molecular docking and dynamics simulation studies. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2021.1942217
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19(14):1639–1662
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791
Murray MY, Rushworth SA, Zaitseva L, Bowles KM, Macewan DJ (2013) Attenuation of dexamethasone-induced cell death in multiple myeloma is mediated by miR-125b expression. Cell Cycle 12:2144–2153
Pelicano H, Martin DS, Xu R-H, Huang P (2006a) Glycolysis inhibition for anticancer treatment. Oncogene 25:4633–4646. https://doi.org/10.1038/sj.onc.1209597
Pelicano H, Martin DS, Xu RH, Huang P (2006b) Glycolysis inhibition for anticancer treatment. Oncogene 25(34):4633–4646
Petrelli A, Giordano S (2008) From single- to multi-target drugs in cancer therapy: when aspecificity becomes an advantage. Curr Med Chem 15:422–432
RCSB Protein Data Bank ÐRCSB PDB (2016) [Internet]. [cited 28 Nov 2016]. http://www.rcsb.org/pdb/home/home.do
Robey RB, Hay N (2006) Mitochondrial hexokinases, novel mediators of the antiapoptotic effects of growth factors and Akt. Oncogene 25(34):4683–4696
Roy A, Bhatia KS (2020) In silico analysis of plumbagin against cyclin-dependent kinases receptor. Vegetos 34:50–56. https://doi.org/10.1007/s42535-020-00169-8
Suda K, Mitsudomi T (2014) Successes and limitations of targeted cancer therapy in lung cancer. Prog Tumor Res 41:62–77. https://doi.org/10.1159/000355902
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc.21334
Vander Heiden MG (2011) Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov 10:671–684. https://doi.org/10.1038/nrd3504
Vander Heiden MG, Cantley LC, Thompson CB (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324:1029–1033. https://doi.org/10.1126/science.1160809
Verma AK, Hossain MS, Ahmed SF, Hussain N, Ashid M, Upadhyay SK, Srivastava SK (2022) In silico identification of ethoxy phthalimide pyrazole derivatives as IL-17A and IL-18 targeted gouty arthritis agents. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2022.2071338
Vijayakumar S, Rajalakshmi S (2021) Exploring novel natural compound inhibitors for Parkinsonian receptor (DJ1) by homology modeling, molecular docking and MD simulations. Vegetos 34(4):959–970. https://doi.org/10.1007/s42535-021-00263-5
Wang H (2004) Pretreatment with dexamethasone increases antitumor activity of carboplatin and gemcitabine in mice bearing human cancer xenografts: in vivo activity, pharmacokinetics, and clinical implications for cancer chemotherapy. Clin Cancer Res 10:1633–1644
Wang H, Wang Y, Rayburn E, Hill D, Rinehart J, Zhang R (2007) Dexamethasone as a chemosensitizer for breast cancer chemotherapy: potentiation of the antitumor activity of adriamycin, modulation of cytokine expression, and pharmacokinetics. Int J Oncol 30:947–953
Warburg O (1956) On the origin of cancer cells. Science 123(3191):309–314
Wilson JE (2003) Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. J Exp Biol 206(12):2049–2057
Witschi H, Espiritu I, Ly M, Uyeminami D (2005) The chemopreventive effects of orally aDMSinistered dexamethasone in strain A/J mice following cessation of smoke exposure. Inhal Toxicol 17:119–122
Yadav S, Pandey SK, Singh VK, Goel Y, Kumar A, Singh SM (2017) Molecular docking studies of 3-bromopyruvate and its derivatives to metabolic regulatory enzymes: Implication in designing of novel anticancer therapeutic strategies. PLoS ONE 12(5):e0176403. https://doi.org/10.1371/journal.pone.0176403
Yano A, Fujii Y, Iwai A, Kageyama Y, Kihara K (2006) 433: Glucocorticoids suppress tumor angiogenesis and in vivo growth of prostate cancer cells. J Urol 175:141
Acknowledgements
The authors acknowledge the support of Multiscale Simulation Research Center (MSRC), Manipal University Jaipur for computational work. AKV acknowledges Dr. Ramdas Pai fellowship from Manipal University Jaipur.
Author Information
Structural Biology & Bioinformatics Laboratory, Department of Biosciences, Manipal University Jaipur, Jaipur, India
Structural Biology & Bioinformatics Laboratory, Department of Biosciences, Manipal University Jaipur, Jaipur, India
sandeepkumar.srivastava@jaipur