Amplified fragment length polymorphism analysis to assess genetic diversity in rice
Research Articles | Published: 02 December, 2019
First Page: 83
Last Page: 91
Views: 3820
Keywords: Genetic diversity, Rice, AFLP, Productive lines, Superior genetics
Abstract
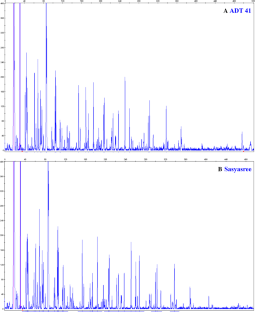
Amplified Fragment Length polymorphism (AFLP) was studied involving 48 diverse rice genotypes, comprising of indica, japonica, and Tongil type (indica × japonica) varieties to assess the genetic diversity. Initially 64 primer pair combinations were screened of which 5 were found to be suitable producing clear fragments (35–500 bp). In total 700 amplicons (bands) were produced with an average of 140 bands per primer pair. Maximum (207) from EcoRI AC*/MseI-CAC and minimum (75) amplicons from EcoRI TG*/MseI-CTT primer pairs were observed. Minimum (0.973) and maximum (0.990) PIC values recorded for primer pair EcoRI AC*/MseI-CAC and EcoRI AC*/MseI-CAT, respectively. Pairwise genetic similarity estimates ranged in between 0.6412 and 0.943 with a mean of 0.797. Triguna, an indica high yielding variety (HYV), and a new plant type (NPT) rice, IR 7946-46-1-3-2 having indica x japonica parentage showed minimum genetic similarity coefficient (0.641) with maximum genetic divergence, whereas ADT 41 and Sasyasree, two indica HYVs displayed maximum genetic similarity coefficient—0.953 with lowest genetic distance. All accessions could be clearly identified by using five primer pairs, offers immense scope of AFLP in assessing genetic diversity in rice. The present study also identified prospective varieties for use in selective hybridization and productive progeny selection. Most of the varieties shared distinct clusters based on varietal types, race, parentage and growing area where those are cultivated traditionally albeit with a few exceptions. The dendrogram could demarcate the varieties irrespective of race with unique traits, albeit with exceptions, which warrants further experimentation involving more number of primers in future.
References
- Anjana B, Choudhury PR, Pande V, Mandal AB (2015) Variety identification and genetic diversity analysis in rice (Oryza sativa L.) using STMS markers. Int J Fundam Appl Sci 4(3):72–80
- Anon (2014) All India Area, Production and Yield of Rice along with Coverage under Irrigation, Agriculture Statistics at a Glance 2014, Department of Agriculture and Cooperation, Directorate of Economics and Statistics. Govt of India, New Delhi, pp 452
- Barrett BA, Kidwell KK (1998) AFLP-based genetic diversity assessment amongwheat cultivars from the Pacific Northwest. Crop Sci 38:1261–1271
- Bates SRE, Knorr DA, Weller JW, Ziegle JS (1996) Instrumentation for automated molecular marker acquisition and data analysis. In: The Impact of plant molecular genetics. pp 239–255
- Brubaker CL, Wendel JF (1994) Reevaluating the origin of domesticated cotton (Gossypium hirsutum; Malvaceae) using nuclear restriction fragment length polymorphisms (RFLPs). Am J Bot 81(10):1309–1326
- Burkhamer RL, Lanning SP, Martens RJ, Martin JM, Talbert LE (1998) Predicting progeny variance from parental divergence in hard red spring wheat. Crop Sci. 38:243–248
- Cheung WY, Moore G, Money TA, Gale MD (1992) HpaII library indicates‘methylation-free islands’ in wheat and barley. Theor Appl Genet 84:739–746
- Cho YG, Blair MW, Panaud O, McCouch SR (1996) Cloning and mapping of variety specific rice genomic DNA sequences: amplified fragment-length polymorphism (AFLP) from silver stained polyacrylamide gels. Genome 39:373–378
- Cho YC, Shin YS, Ahn SN, Gleen BG, Kang KH, Darshan B, Moon HP (1999) DNA fingerprinting of rice cultivars using AFLP and RAPD markers. Korean J Crop Sci 44(1):26–31
- Conway G, Toenniessen G (1999) Feeding the world in the twenty-first century. Nature 402(6761):C55
- Das M, Banerjee S, Topdar N, Kundu A, Sarkar D, Sinha MK, Balyan HS, Gupta PK (2011) Development of large-scale AFLP markers in jute. J Plant Biochem Biotechnol 20(2):270–275
- Datta SK (2002) Recent developments in transgenics for abiotic stress tolerance in rice. JIRCAS Work Rep 2002:43–53
- Hill M, Witsenboer H, Zabeau M, Vos P, Kesseli R, Michelmore R (1996) PCR-based fingerprinting using AFLPs as a tool for studying genetic relationships in Lactuca spp. Theor Appl Genet 93(8):1202–1210
- Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat 44:223–270
- Khush GS, Baenziger PS (1998) Crop improvement: emerging trends in rice and wheat. In: Crop Productivity and Sustainability—Shaping the Future, Proceedings of the 2nd International Crop Science Congress, pp 113–125)
- Lookhart GEOR, Bean S (1995) Separation and characterization of wheat protein fractions by high-performance capillary electrophoresis. Cereal Chem 72(6):527–532
- Mackill DJ, Zhang Z, Redona ED, Colowit PM (1996) Level of polymorphism and genetic mapping of AFLP markers in rice. Genome 39(5):969–977
- Manifesto MM, Schlatter AR, Hopp HE, Suárez EY, Dubcovsky J (2001) Quantitative evaluation of genetic diversity in wheat germplasm using molecular markers. Crop Sci 41(3):682–690
- Martos V, Royo C, Rharrabti Y, Del Moral LG (2005) Using AFLPs to determine phylogenetic relationships and genetic erosion in durum wheat cultivars released in Italy and Spain throughout the 20th century. Field Crops Res 91(1):107–116
- Messmer MM, Melchinger AE, Hermann RG, Boppermaier J (1993) Relationships among early European Karakousis A, Kretschmer JM, Manning S, Langridge P (1998) Genetic diversity in Australian wheat varietes and breeding material based on RFLP data. Theor Appl Genet 96:435–446
- Metakovsky EV, Branlard G (1998) Genetic diversity of French common wheat germplasm based on gliadin alleles. Theor Appl Genet 96(2):209–218
- Murtaza N (2006) Cotton genetic diversity study by AFLP markers. Electron J Biotechnol 9(4)
- Peng S, Tang Q, Zou Y (2009) Current status and challenges of rice production in China. Plant Prod Sci 12(1):3–8
- Powel W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2(3):225–238
- Subudhi PK, Nandi S, Casal C, Virmani SS, Huang N (1998) Classification of rice germplasm: III. High-resolution fingerprinting of cytoplasmic genetic male-sterile (CMS) lines with AFLP. Theor Appl Genet 96(6–7):941–949
- Varshney RK, Bansal KC, Agarwal PK, Datta SK, Craufurd PQ (2011) Agricultural biotechnology for crop improvement in variable climate: hope or hype? Trends Plant Sci 16:363–371
- Vkuylstee M, Mank R, Antonise R, Bastiaans E, Senior ML, Stuber CW, Melchinger AE, Lübberstedt T, Xxia C, Stam P, Zabeau M, Kuiper M (1999) Two high-density AFLP® linkage maps of Zea mays L.: analysis of distribution of AFLP markers. Theor Appl Genet 99:921–935
- Vos P, Hogers R, Bleeker M, Reijans M, Van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
- Wang ZY, Second G, Tanksley SD (1992) Polymorphism and phylogenetic relationships among species in the genus Oryza as determined by analysis of nuclear RFLPs. Theor Appl Genet 83(5):565–581
- Zhu J, Gale MD, Quarrie S, Jackson MT, Bryan GJ (1998) AFLP markers for the study of rice biodiversity. Theor Appl Genet 96(5):602–611
Author Information
ICAR-Indian Institute of Seed Science, Mau, India