Assessment of genetic diversity of upland rice (Oryza sativa L.) genotypes from North Eastern Hill Region of India
Syiemlieh Paharasainiing, Noren S. K., Khanna V. K., Rai Mayank, Meetei N. T., Pattanayak A.
Research Articles | Published: 01 July, 2019
First Page: 407
Last Page: 419
Views: 3619
Keywords: Rice, SSR markers, North Eastern Hills Region
Abstract
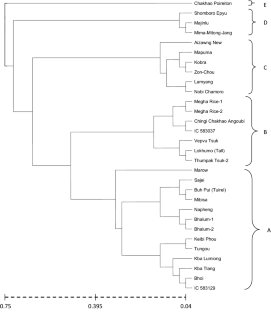
Genetic diversity among 30 upland rice genotypes (Oryza sativa L.) was evaluated using morphological and SSR markers. Roger’s distance was calculated for all the data sets as a measure of genetic distance between accessions. The distance matrices were used to generate a dendrogram based on the UPGMA method. The minimum genetic distance based on morphological marker was 0.04 which was found between Megha Rice-1 and Megha Rice-2. The maximum genetic distance was 0.75 observed between Aizawng New and Majinlu, Aizawng New and Mima-Mitong-Jang, Mibisa and Chakhao Poireiton, Chakhao Poireiton and Vepvu Tsuk with an average of 0.395. The number of alleles per SSR marker varied from 2 to 8, with an average number of 4.286 alleles per locus. Polymorphism information content (PIC) values ranged from 0.062 to 0.799 per locus with an average of 0.425. The minimum and maximum genetic distances based on Roger’s distance, were found to be 0.00 (Bhalum-1 and Bhalum-2) and 0.84 (Aizawng New and Mibisa), respectively. In cluster analysis, morphological and molecular marker-based genetic distance grouped the local landraces together with the improved lines and Gene Bank lines. Correlation analysis between morphological data and molecular data revealed a very poor association (r = 0.051 at p = 0.271). The diverse genotypes revealed from morphological-based dendrogram were the following pairs viz., Aizawng New and Majinlu, Aizawng New and Mima-Mitong-Jang, Mibisa and Chakhao Poireiton, Chakhao Poireiton and Vepvu Tsuk, Buh Pui (Tuirel) and Chakhao Poireiton, Marow and Chakhao Poireiton, Kba Tlang and Majinlu, Lokhumo (Tall) and Chakhao Poireiton. While Aizawng New and Mibisa, IC 583129 and Mibisa, Bhalum-1 and Mibisa, Bhalum-2 and Mibisa, Marow and Mibisa pairs were identified as the diverse genotypes based on molecular dendrogram which may be used as parent for hybridization during further rice improvement.
References
- Akagi H, Yokozeki Y, Inagaki A, Fujimura T (1996) Microsatellite DNA markers for rice chromosomes. Theor Appl Genet 93:1071–1077
- Anderson JC, Autrique J, Tanksley S, Sorrells M (1993) Optimizing parent selection for genetic linkage maps. Genome 36:181–186
- Ashfaq M, Khan AS (2012) Genetic diversity in Basmati rice (Oryza sativa L.) germplasm as revealed by microsatellite (SSR) markers. Russ J Genet 48(1):53–62
- Chuang H, Lur H, Hwu K, Chang M (2011) Authentication of domestic Taiwan rice varieties based on fingerprinting analysis of microsatellite DNA markers. Bot Stud 52:393–405
- Guei GR (2000) Participatory varietal selection and rice biodiversity at community levels. In: Paper presented at the participatory varietal selection workshop, West Africa Rice Development Association, pp 13–21
- Gupta S (1990) Physico-chemical characters of some rice cultivars of West Bengal. Int Rice Res Newsl 15(1):12
- Khush GS (2005) What it will take to feed five billion rice consumers by 2030. Plant Mol Biol 59:1–6
- Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
- McCouch SR, Chen X, Panaud O, Temnykh S, Xu Y, Cho YG, Huang N, Ishii T, Blair M (1997) Microsatellite marker development, mapping and applications in rice genetics and breeding. Plant Mol Biol 35(1–2):89–99
- McCouch SR, Teytelman L, Xu Y, Lobos KB, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, Zhang Q, Kono I, Yano M, Fjellstrom R, DeClerk G, Schneider D, Cartinhour S, Ware D, Stein L (2002) Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res 9:199–207
- Pessoa-Filho M, Belo A, Alcochete AAN, Rangel PHN, Ferreira ME (2007) A set of multiplex panels of microsatellite markers for rapid molecular characterization of rice accessions. BMC Plant Biol 7:23
- Rabbani MA, Masood MS, Shinwari ZK, Yamaguchi-Shinozaki K (2010) Genetic analysis of Basmati and non-Basmati Pakistani rice (Oryza sativa L.) cultivars using microsatellite markers. Pak J Bot 42(4):2551–2564
- Rahman MS, Sohag MKH, Rahman L (2010) Microsatellite based DNA fingerprinting of 28 local rice (Oryza sativa L.) varieties of Bangladesh. J Bangladesh Agric Univ 8(1):7–17
- Ram K, Chetty CVS (1934) Classification of rice of Bihar and Orissa. Indian J Agric Sci 4:537
- Ram K, Ekbote RB (1936) Classification of the Punjab and Western United Provinces of wheat. Indian J Agric Sci 6:930–937
- Rogers JS (1972) Measures of genetic similarity and genetic distance. Studies in genetics, vol 7. University of Texas, Austin, pp 145–153
- Sarma MK, Richharia AK, Agarwal RK (2004) Characterisation of ahu rice of Assam for morphological and agronomic traits under transplanted conditions. Oryza 41(1–2):8–12
- Sethi RL, Saxena BP (1930) Classification and study of characters of cultivated rice in the United Provinces. Mem Dept Agric India Bot Ser 18:140–209
- Shah R, Sulemani MZ, Balah MS, Hassan G (1999) Performance of coarse rice genotypes in plains of DI Khan, Pakistan. Pak J Biol Sci 2:507–509
- Sharma KK, Ahmed T, Baruah DK (1990) Grain characteristics of some aromatic rice varieties of Assam. Int Rice Res Newsl 15(1):13
- Siddiq EA, Ahmed MI, Viraktamath BC, Rangasamy M, Vijayakumar R, Vidyachandra B, Zaman FU, Chatterjee SD (1998) Hybrid rice technology in India: current status and future outlook. In: Virmani SS, Siddiq EA, Muralidharan K (eds) Advances in hybrid rice technology. International Rice Research Institute, Manila, pp 311–324
- Smith JSC, Smith OS (1992) Fingerprinting crop varieties. Adv Agron 47:85–140
- Smykal P, Horacek J, Dostalova R, Hybl M (2008) Variety discrimination in pea (Pisum sativum L.) by molecular, biochemical and morphological markers. J Appl Genet 49:155–166
- Vanniarajan C, Vinod KK, Pereira A (2012) Molecular evaluation of genetic diversity and association studies in rice (Oryza sativa L.). J Genet 91(1):9–19
- Weir BS (1990) Genetic data analysis: methods for discrete population genetic data. Sinauer Associates, Inc., Sunderland, p 377
- Xie J, Agrama HA, Kong D, Zhuang J, Hu B, Wan Y, Yan W (2010) Genetic diversity associated with conservation of endangered Dongxiang wild rice (Oryza rufipogon). Genet Resour Crop Evol 57:597–609
- Zafar N, Aziz S, Masood S (2004) Phenotypic divergence for agro-morphological traits among landrace genotypes of rice (Oryza sativa L.) from Pakistan. Int J Agric Biol 6(2):335–339
- Holsinger KE, Weir BS (2009) Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat Rev Genet 10(9):639–650
Author Information
School of Crop Improvement, College of Post Graduate Studies in Agricultural Sciences (Central Agricultural University, Imphal), Umiam, India