Chromosomal and PCR-based molecular characterization of Hedychium spp. of Tripura, North-East India
Research Articles | Published: 26 September, 2019
First Page: 521
Last Page: 531
Views: 4219
Keywords: Hedychium spp., ISSR, Karyotype, SSR, UPGMA
Abstract
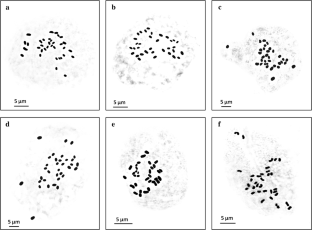
Hedychium is an important genus of Zingiberaceae because of its medicinal and horticultural importance. Investigations on chromosomal and polymerase chain reaction (PCR) based molecular characterization of the genus Hedychium at inter-specific and intra-specific levels have been undertaken for the detailed study of the karyotypes as well as for assessing the genetic diversity. We have analysed chromosomal and molecular marker based characters of three Hedychium spp., viz., H. coccineum, H. coronarium and H. thyrsiforme from the state of Tripura but because of their restricted distributions in this biodiversity hotspot we included only two populations of each species in intra-species analysis. Numerical data of karyotypes reveal that Pop-I and Pop-II of all the three species have the same chromosome count 2n = 34 with a basic number X = 17. In-spite of the close resemblances of their karyotypes, chromosomal data based clustering pattern and their sub-grouping at intra-species level validates the taxonomic status of the species. The inter simple sequence repeat (ISSR) and simple sequence repeat (SSR) dendrogram obtained by unweighted pair group method with arithmetic mean (UPGMA) analysis depicts a local position effect based clustering pattern with intra-specific diversity. Our results also indicate that SSR marker is more reliable in assessing the genetic relationship between and within the species of Hedychium. As Gower’s similarity coefficient is an indicator of cryptic changes in chromosome structure, we propose that dissimilarity in mean centromeric asymmetry (MCA) value at inter-specific level can be related with molecular markers based genetic diversity having high degree of polymorphism.
References
- Basak S, Ramesh AM, Kesari V, Parida A, Mitra S, Rangan L (2014) Genetic diversity relationship of Hedychium from Northeast India as dissected using PCA analysis and hierarchical clustering. Meta Gene 2:459–468. https://doi.org/10.1016/j.mgene.2014.05.002
- Bisht GS, Awasthi AK, Dhole TN (2006) Antimicrobial activity of Hedychium spicatum. Fitoterapia 21:439–443. https://doi.org/10.1016/j.fitote.2006.02.004
- Bornet B, Branchard M (2004) Use of ISSR fingerprints to detect microsatellites and genetic diversity in several related Brassica taxa and Arabidopsis thaliana. Hereditas 140:245–248. https://doi.org/10.1111/j.1601-5223.2004.01737.x
- Chopra RN, Chopra IC, Nayar SL (1956) Glossary of Indian medicinal plants. Council of Scientific and Industrial Research, New Delhi
- Das A, Kesari V, Satyanarayana VM, Parida A, Rangan L (2011) Genetic relationship of Curcuma species from Northeast India using PCR-based markers. Mol Biotechnol 49:65–76. https://doi.org/10.1007/s12033-011-9379-5
- Datta M, Agarwal B (1992) Intervarietal differences in karyotype of tea. Cytologia 57:437–441. https://doi.org/10.1508/cytologia.57.437
- Deb DB (1981) The flora of Tripura state, vol I. Today and Tomorrow’s Publishers, New Delhi
- Deb DB (1983) The flora of Tripura state, vol II. Today and tomorrow’s publishers, New Delhi
- Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26:297–302. https://doi.org/10.2307/1932409
- Gao L, Liu N, Huang B, Hu X (2008) Phylogenetic analysis and genetic mapping of Chinese Hedychium using SRAP markers. Sci Hort 117:369–377. https://doi.org/10.1016/j.scienta.2008.05.016
- Gopanraj G, Dan M, Shiburaj S, Sethuraman MG, George V (2005) Chemical composition and antibacterial activity of the rhizome oil of Hedychium larsenii. Acta Pharm 55:315–320
- Gower JC (1971) A general coefficient of similarity and some of its properties. Biometrics 27:857–871. https://doi.org/10.2307/2528823
- Hammer Ø, Harper DAT, Ryan PD (2001) PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electronica 4:9. http://palaeo-electronica.org/2001_1/past/issue1_01.htm
- Huziwara Y (1962) Karyotype analysis in some genera of compositae. VIII. Further studies on the chromosome of Aster. Am J Bot 49:116–119. https://doi.org/10.1002/j.1537-2197.1962.tb14916.x
- Jadhav V, Kore A, Kadam VJ (2007) In-vitro pediculicidal activity of Hedychium spicatum essential oil. Fitoterapia 78:470–473. https://doi.org/10.1016/j.fitote.2007.02.016
- Jatoi SA, Kikuchi A, Watanabe KN (2007) Genetic diversity, cytology, and systematic and phylogenetic studies in Zingiberaceae. Genes Genom Genet 1:56–62
- Joseph R, Joseph T, Jose J (1999) Karyomorphological studies in the genus Curcuma Linn. Cytologia 64:313–317. https://doi.org/10.1508/cytologia.64.313
- Joy B, Rajan A, Abraham E (2007) Antimicrobial activity and chemical composition of essential oil from Hedychium coronarium. Phutother Res 21:439–443. https://doi.org/10.1002/ptr.2091
- Kantety RV, Zeng X, Bebbetzen JL, Zehr BE (1995) Assessment of genetic diversity in dent and popcorn (Zea mays L.) inbreed lines using inter-simple sequence repeat (ISSR) amplification. Mol Breed 1:36–373. https://doi.org/10.1007/BF01248414
- Kimura M, Crow JF (1964) The number of alleles that can be maintained in a finite population. Genetics 49:725–738
- Kress WJ, Prince LM, Williams KJ (2002) The phylogeny and a new classification of the gingers (Zingiberaceae): evidence from molecular data. Am J Bot 89:1682–1696. https://doi.org/10.3732/ajb.89.10.1682
- Larsen K, Smith RM (1978) A NEW species of Curcuma from Thailand. Notes R Bot Garden Edinburgh 30:269–272
- Levan A, Fredga K, Sandbery AA (1964) Nomenclature for centromeric position on chromosome. Heriditas 52:201–220. https://doi.org/10.1111/j.1601-5223.1964.tb01953.x
- Lewontin RC (1972) Testing the theory of natural selection. Nature 236:181–182. https://doi.org/10.1038/236181a0
- Mahanty HK (1963) A Karyologcal study of certain Zingiberaceae with reference to their taxonomy. M.Sc. Thesis, University of London, UK
- Mantel N (1967) The detection of disease clustering and generalized regression approach. Cancer Res 27:209–220
- Mao AA, Hynniewta TM, Sanjappa M (2009) Plant wealth of northeast India with reference to ethnobotany. Indian J Tradit Knowl 8:96–103
- Medeiros JR, Campos LB, Mendonca SC, Davin LB, Lewis NG (2003) Composition and antimicrobial activity of the essential oils from invasive species of the Azores, Hedychium gardnerianum and Pittosporum undulatum. Phytochemistry 64:561–565. https://doi.org/10.1016/S0031-9422(03)00338-8
- Mukherjee I (1970) Chromosome studies of some species of Hedychium. Bot Mag 83:237–241
- Myers N, Mittermier RA, Mittermier CG, Fonseca GA, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 40:853–858. https://doi.org/10.1038/35002501
- Naithani S, Sarbhoy R (1973) Cytological studies in Lens esculenta Moench. Cytologia 38:195–203. https://doi.org/10.1508/cytologia.38.195
- Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323. https://doi.org/10.1073/pnas.70.12.3321
- Parida R, Mohanty S, Nayak S (2013) In vitro propagation of Hedychium coronarium Koen. through axillary bud proliferation. Plant Biosystems 147:905–912. https://doi.org/10.1080/11263504.2012.748102
- Peruzzi L, Eroğlu HE (2013) Karyotype asymmetry: again, how to measure and what to measure? Comp Cytogenet 7:1–9. https://doi.org/10.3897/CompCytogen.v7i1.4431
- Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238. https://doi.org/10.1007/BF00564200
- Rajkumari S, Sanatombi K (2018) Genetic diversity analysis of Hedychium species based on RAPD and ISSR markers. Proc Natl A Sci India B. https://doi.org/10.1007/s40011-018-0976-y
- Rohlf FJ (2000) NTSYSpc: numerical taxonomy system. version 2.21c. Exeter Software, Setauket
- Roldan IR, Dendauw J, Van EB, Depicker A, Loose MD (2000) AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol Breed 6:125–134. https://doi.org/10.1023/A:1009680614564
- Saleh MM, Kamal RM, Abdulla WA (1982) A contribution to the molluscicidal activity of Hedychium gardinarum. Planta Med 45:166. https://doi.org/10.1055/s-2007-971366
- Sharma AK, Bhattacharyya NK (1959) Cytology of Several members of Zingiberaceae. La cellule 59:297–346
- Sharma AK, Sharma A (1980) Chromosome technique theory and practical, 3rd edn. Butter works Ltd., London
- Shrotriya S, Ali MS, Saha A, Bachar SC, Islam MS (2007) Anti-inflammatory and analgesic effects of Hedychium coronarium Koen. Pak J Pharm Sci 20:47–51
- Sirirugsa P, Larsen K (1995) The genus Hedychium (Zingiberaceae) in Thailand. Nord J Bot 15:301–304. https://doi.org/10.1111/j.1756-1051.1995.tb00156.x
- Sneath PHA, Sokal RR (1973) Numerical taxonomy: the principles and practice of numerical classification. W. H. Freeman and company, San Francisco
- Sokal RR, Michener CD (1958) A statistical method for evaluating systematic relationships. Sci Bull 38:1409–1438
- Stebbins GL (1971) Chromosomal evolution in higher plants. Edward Arnold Ltd., London
- Thomas S, Mani B, Britto SJ (2015) A new species of Hedychium (Zingiberaceae) from the southern Western Ghats, India. Webbia 70:221–225. https://doi.org/10.1080/00837792.2015.1082270
- Warren KS, Peters PA (1968) Cercariae of Schistosoma mansoni and plants: attempt to penetrate Phaseolus vulgaris and Hedychium coronarium produces a cercaricide. Nature 217:647–648. https://doi.org/10.1038/217647a0
- Watanabe K, Yahara T, Denda T, Kosuge K (1999) Chromosomal evolution in the genus Brachyscome (Asteraceae, Astereae): statistical tests regarding correlation between changes in karyotype and habit using phylogenetic information. J Plant Res 112:145–161. https://doi.org/10.1007/PL00013869
- Yeh FC, Yang RC, Boyle TBJ, Ye ZH, Mao JX (1997) POPGENE, the user friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Centre, University of Alberta, Alberta
- Yu F, Kress WJ, Gao JY (2010) Morphology, distribution, and chromosome counts of two varieties of Hedychium villosum (Zingiberaceae). J Syst Evol 48:344–349. https://doi.org/10.1111/j.1759-6831.2010.00094.x
- Zarco CR (1986) A new method for estimating karyotype asymmetry. Taxon 35:526–530. https://doi.org/10.2307/1221906
- Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176–183. https://doi.org/10.1006/geno.1994.1151
Author Information
Cytogenetics and Plant Biotechnology Laboratory, Department of Botany, Tripura University, Agartala, India