Estimation of genetic diversity for interspecific hybridization in Pleurotus spp
Barh Anupam, Sharma V. P., Annepu Sudheer Kumar, Kumari Babita, Kamal Shwet, Kumar Anil
Research Articles | Published: 29 January, 2023
First Page: 155
Last Page: 164
Views: 3412
Keywords: Oyster mushroom, ITS, ISSR, Genetic improvement, Mahalanobis distance, Transgressive segregants
Abstract
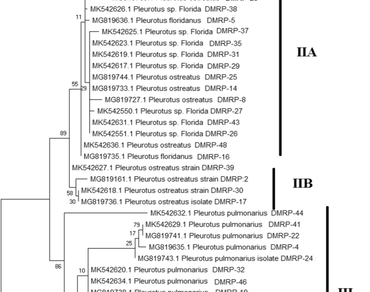
Pleurotus spp. is a taxonomically diverse group of species having a large range of distribution. The trait enhancement for commercial exploitation can be possible using inter and intraspecific hybridization in consideration of mating behaviour. In our study, we used forty-eight strains of the Pleurotus spp. to select the parents for interspecific hybridization. Diversity was selected as the criteria for selection and was estimated at the molecular and phenotypic levels. Nine inter-simple sequence repeat (ISSR) markers were used for the estimation of molecular diversity. All 48 strains were segregated into four species based on ITS maker (internal transcribed spacer) molecular identification. ISSR marker-based Unweighted Pair-Group Method with Arithmetic Mean (UPGMA) dendrogram was created, and diverse strains were identified. The phenotypic genetic divergence based on quantitative characters was also computed using Mahalanobis D2 statistics. Based on the genetic distance from polymorphism information content, mahalanobis distance, and mean of biological efficiency, the parental combinations of DMRP-10 × DMRP-45, DMRP-25 × DMRP-36, DMRP-8 × DMRP-25, DMRP-25 × DMRP-41, and DMRP-10 × DMRP-25 were identified for further hybridization.
References
Albani MC, Battey NH, Wilkinson MJ (2004) The development of ISSR-derived SCAR markers around the Seasonal flowering locus (SFL) in Fragaria vesca. Theor Appl Genet 109:571–579. https://doi.org/10.1007/s00122-004-1654-4
Cohen L, Persky Y, Hadar R, Persky L, Hadar Y (2002) Biotechnological applications and potential of wood-degrading mushrooms of the genus Pleurotus. Appl Microbiol Biotechnol 58:582–594. https://doi.org/10.1007/s00253-002-0930-y
Das S, Deb B (2015) DNA barcoding of fungi using ribosomal ITS marker for genetic diversity analysis: a review. Int J Pure Appl Biosci 3:160–167
Gonzalez P, Labarère J (2000) Phylogenetic relationships of Pleurotus species according to the sequence and secondary structure of the mitochondrial small-subunit rRNA V4, V6 and V9 domains. Microbiology 146:209–221. https://doi.org/10.1099/00221287-146-1-209
Guzman G (2000) Genus Pleurotus (Jacq.: Fr.) P. Kumm. (Agaricomycetideae): diversity, taxonomic problems, and cultural and traditional medicinal uses. Int J Med Mushrooms 2:29. https://doi.org/10.1615/IntJMedMushr.v2.i2.10
He L, Yao K, Xu X et al (2022) Intraspecific identification of lingzhi medicinal mushroom, Ganoderma lingzhi (Agaricomycetes), using ISSR markers. Int J Med Mushrooms 24:43–52. https://doi.org/10.1615/INTJMEDMUSHROOMS.2022043141
Joshi M, Verma SK, Singh JP, Barh A (2013) Genetic diversity assessment in Lentil (Lens culinaris Medikus) genotypes through ISSR marker. The Bioscan 8:1529–1532
Lallawmsanga PAK, Mishra VK et al (2016) Antimicrobial potential, identification and phylogenetic affiliation of wild mushrooms from two sub-tropical semi-evergreen indian forest ecosystems. PLoS ONE 11:e0166368. https://doi.org/10.1371/journal.pone.0166368
Liu J, Wang Z-R, Li C et al (2015) Evaluating genetic diversity and constructing core collections of Chinese Lentinula edodes cultivars using ISSR and SRAP markers. J Basic Microbiol 55:749–760. https://doi.org/10.1002/jobm.201400774
Menolli Junior N, Asai T, Capelari M, Paccola-Meirelles LD (2010) Morphological and molecular identification of four Brazilian commercial isolates of Pleurotus spp. and cultivation on corncob. Brazilian Arch Biol Technol 53:397–408. https://doi.org/10.1590/S1516-89132010000200019
Nazrul MI, YinBing B (2011) Differentiation of homokaryons and heterokaryons of Agaricus bisporus with inter-simple sequence repeat markers. Microbiol Res 166:226–236. https://doi.org/10.1016/J.MICRES.2010.03.001
Rao CR (1952) Advance statistical methods in biometrical research. John Wiley and Sons Inc., New York
Rohlf FJ (2008) NTSYSpc: numerical taxonomy system, ver. 2.20.
Royse BJ, Tan Q (2017) Current overview of mushroom production in the world. In: Zied DC, Giménez, Arturo P (eds) Edible and medicinal mushrooms: technology and applications, 1st edn. John Wiley & Sons Ltd, New York, pp 5–13
Shnyreva AA, Shnyreva AV (2015) Phylogenetic analysis of Pleurotus species. Genetika 51:177–187
Su H, Wang L, Liu L et al (2008) Use of inter-simple sequence repeat markers to develop strain-specific SCAR markers for Flammulina velutipes. J Appl Genet 49:233–235. https://doi.org/10.1007/BF03195619
Sudhir K, Glen S, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Tang L, Xiao Y, Li L et al (2010) Analysis of genetic diversity among Chinese auricularia auricula cultivars using combined ISSR and SRAP markers. Curr Microbiol 61:132–140. https://doi.org/10.1007/s00284-010-9587-4
Upadhyay R, Verma B, Sood S et al (2017) Documentary of agaricomycetous mushrooms of India orders—agaricales, boletales and russulales. JayaPublishing House, Delhi
Zhao M, Huang C, Chen Q et al (2013) Genetic variability and population structure of the mushroom Pleurotus eryngii var. tuoliensis. PLoS ONE 8:e83253. https://doi.org/10.1371/journal.pone.0083253
Author Information
ICAR-Directorate of Mushroom Research, Solan, India