Evaluation of amplification potential of species identification markers in a virtual PCR environment and efficient molecular identification of three black truffle species by multiplex PCR assay
*Article not assigned to an issue yet
Song Ju-Il, Choe Mi-Ryong, Ho Un-Hyang, Ri Sok-Jun, Kim Mi-Hyang, Ri Tae-Ryong, Yo Chung-Il, Kim Yong-Nam
Research Articles | Published: 10 November, 2025
First Page: 0
Last Page: 0
Views: 96
Keywords: Tuber, Virtual PCR environment, Multiplex PCR, DNA barcode, In silico
Abstract
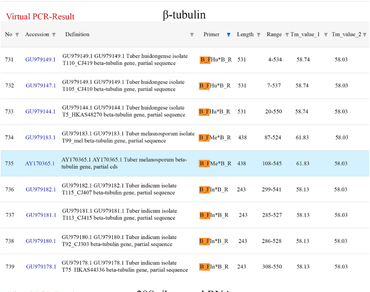
There has been a great deal of research in the field of artificial cultivation of black truffles including Tuber huidongense, Tuber melanosporum and Tuber indicum. Here, molecular real time identification of culture at low cost is very important to ensure quality of truffle products. Identification of truffle species based on genus or species-specific primers developed so far has limitations of being laborious and costly, as sequencing or PCR are required. In this study, we constructed in silico virtual PCR environment being able to confirm amplification potential of species-specific markers and conducted molecular discrimination of 3 truffle species by multiplex PCR. Evaluation of amplification potential in the virtual PCR environment and multiplex PCR assay using DNA barcode genes (ITS, β-tubulin, and 28S rRNA) would contribute to molecular species identification of truffle species in the future.
References
Amicucci A, Guidi C, Zambonelli A, Potenza L, Stocchi V (2000) Multiplex PCR for the identification of white Tuber species. FEMS Microbiol Lett 189:265–269
Bonito G, Smith ME, Nowak M, Healy RA, Guevara G, Cazares E, Kinoshita A, Nouhra ER, Domı´nguez LS, Tedersoo L, Murat C, Wang Y, Moreno BA, Pfister DH, Nara K, Zambonelli A, Trappe JM, Vilgalys R (2013) Historical biogeography and diversification of truffles in the Tuberaceae and their newly identified southern hemisphere sister lineage. PLoS ONE 8(1):e52765
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Francesco Paolocci A, Andrea Rubini A, Bruno Granetti A, Sergio Arcioni A (1999) Rapid molecular approach for a reliable identification of Tuber spp. ectomycorrhizae. FEMS Microbiol Ecol 28:23–30
Glass NL, Donaldson GC (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 61:1323–1330
Graziosi S, Hall IR, Zambonelli A (2022) The mysteries of the white truffle: its biology. Ecol Cultivat Encyclopedia 2:1959–1971. https://doi.org/10.3390/encyclopedia2040135
Gryndler M, Hrselova H, Soukupova L, Streiblova E, Valda S, Borovicka J, Gryndlerova H, Gazo J, Miko M (2011) Detection of summer truffle (Tuber aestivum Vittad) in ectomycorrhizae and in soil using specific primers. FEMS Microbiol Lett 318:84–91
Ho U-H, Pak S-H, Kim K, Pak H-S (2023) Efficient screening of SNP in canine OR52N9 and OR9S25 as assistant marker of olfactory ability. J Vet Behav 60:51–55
Iott M, Piattoni F, Leonardi P, Hall IR, Zambonelli A (2016) First evidence for truffle production from plants inoculated with mycelial pure cultures. Mycorrhiza. https://doi.org/10.1007/s00572-016-0703-6
Johnson M et al (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36 (Web Server issue): 5–9. https://doi.org/10.1093/nar/gkn201
Kalendar R, Lee D, Schulman AH (2014) FastPCR software for PCR, in silico PCR, and oligonucleotide assembly and analysis. Methods Mol Biol 1116:271–302
Kumar LM, Smith ME, Nouhra ER, Orihara T, Leiva PS, Pfister DH, McLaughlin DJ, Trappe JM, Healy RA (2017) A molecular and morphological reexamination of the generic limits of truffles in the Tarzetta-Geopyxis lineage – Densocarpa, Hydnocystis, and Paurocotylis. Fungal Biol. https://doi.org/10.1016/j.funbio.2016.12.004
Lexa M, Horak J, Brzobohaty B (2001) Virtual PCR. Bioinformatics 17(2):192–193
Luana B, Antonella A, Deborah A, Emanuela P, Lucia P, Chiara G, Vilberto S (1999) A new pair of primers designed for amplification of the ITS region in Tuber species. FEMS Microbiol Lett 173:239–245
Mabru D, Douet JP, Mouton A, Dupre C, Ricard JM, Medina B, Castroviejo M, Chevalier G (2004) PCR-RFLP using a SNP on the mitochondrial Lsu-rDNA as an easy method to differentiate Tuber melanosporum (Perigord truffle) and other truffle species in cans. Int J Food Microbiol 94:33–42
Martínez-Porchas M, Villalpando-Canchola E, Vargas-Albores F (2016) Significant loss of sensitivity and specificity in the taxonomic classification occurs when short 16S rRNA gene sequences are used. Heliyon 2:e00170. https://doi.org/10.1016/j.heliyon.2016.e00170
Murat C, Díez J, Luis P, Delaruelle C, Dupré C, Chevalier G, Bonfante P, Martin F (2004) Polymorphism at the ribosomal DNA ITS and its relation to postglacial re-colonization routes of the Perigord truffle Tuber melanosporum. New Phytol 164:401–411
Riccioni C, Rubini A, Türko˘glu A, Belfiori B, Paolocci F (2019) Ribosomal DNA polymorphisms reveal genetic structure and a phylogeographic pattern in the Burgundy truffle Tuber aestivum Vittad. Mycologia 111:26–39
Rubini A, Paolocci F, Granetti B, Arcioni S (1998) Single step molecular characterization of morphologically similar black truffle species. FEMS Microbiol Lett 164:7–12
Schiaffino A, Lancellotti A, Corda P, Marras F (2006) Discrimination of Tuber aestivum, T. borchii and T. melanosporum by a multiplex PCR. 5th International conference on mycorrhizae, Granada, pp 23–27
Segelke T, Schelm S, Ahlers C, Fischer M (2020) Food authentication: truffle (Tuber spp.) species differentiation by FT-NIR and chemometrics. Foods. https://doi.org/10.3390/foods9070922
Steidinger BS, Büntgen U, Stobbe U, Tegel W, Sproll L, Haeni M, Moser B, Bagi I, Bonet J-A, Buée M, Dauphin B, Peña F, Molinier V, Zweifel R, Egli S, Peter M (2022) The fall of the summer truffle: recurring hot, dry summers result in declining fruitbody production of Tuber aestivum in Central Europe. Glob Change Biol 28:7376–7390
Tang L (2023) Submerged fermentation for truffles: a potential replacement and production. Pharm Bioprocess 11(3):41–43
Ye J et al (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. https://doi.org/10.1186/1471-2105-13-134
Zambonelli A, Iotti M (2006) The pure culture of Tuber mycelia and their use in the cultivation of the truffles. In: Kabar L ed Actes du premier Symposium sur les Champignons hypogés du Bassin Méditerranéen. Rabat, pp 244–255
Zampieri E, Murat C, Cagnasso M, Bonfante P, Mello A (2010) Soil analysis reveals the presence of an extended mycelial network in a Tuber magnatum truffle-ground. FEMS Microbiol Ecol 71:43–49
Author Information
Faculty of Life Science, Kim Il Sung University, Pyongyang, Democratic People’s Republic of Korea