Genetic diversity and population structure of Canna edulis accessions in Vietnam revealed by ISSR markers
Research Articles | Published: 09 May, 2023
First Page: 1009
Last Page: 1018
Views: 4066
Keywords: n Canna edulisn , Conservation, Genetic diversity, ISSR Marker
Abstract
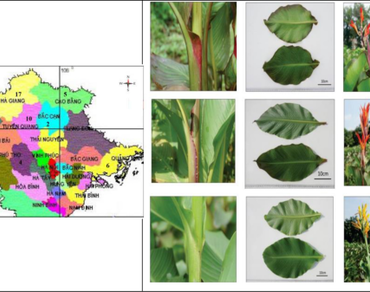
Discovery of the genetic diversity of Canna edulis accessions is valuable for germplasm conservation, improvement, and utilization of this crop as an ornamental garden plant or a food source in the future. This study employed 14 ISSR markers and morphological characters to assess genetic relationships among 52 accessions of Canna edulis (denoted as C1 to C52) collected in 9 provinces of three agroecological regions of Vietnam. A total of 75 loci with an average of 5.2 loci per marker and sizes ranging from 300 to 1900 bp were obtained. Most of the loci were polymorphic with an average polymorphism of 81.88% and a PIC value of 0.735. The Jaccard’s similarity coefficients were in the range of 0.42 and 0.98. The UPGMA analysis grouped the 52 accessions into two groups at the cut-off value of 0.54 with 13 and 39 accessions respectively. Population structure analysis also indicated a similar pattern of grouping for two sub-populations. Genetic variation occured within the population (90%). Additionally, the leaf width and the plant height were the two key morphological traits explaining the differences between the two sub-populations, suggesting for use of these traits in the classification and taxonomy of Canna edulis in Vietnam.
References
Akhi AS, Sarkar BK, Sultana N, Jui ZS, Sarker RH, Rahman O (2021) Molecular characterization of Canna indica L. based on random amplified polymorphic DNA markers. Bangladesh J Plant Taxon 28(1):75–81. https://doi.org/10.3329/bjpt.v28i1.54209
Algar AFC, Umali AB, Tayobong RRP (2019) Physicochemical and functional properties of starch from Philippine edible Canna (Canna indica L.) rhizomes. J Microbiol Biotechnol Food Sci 9(1):34–37. https://doi.org/10.15414/jmbfs.2019.9.1.34-37
Andrade-Mahecha MM, Tapia-Blácido DR, Menegalli FC (2012) Physical-chemical, thermal, and functional properties of achira (Canna indica L.) flour and starch from different geographical origins. Starch/Starke 64(5):348–358. https://doi.org/10.1002/star.201100149
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32(3):314–331
CABI (2020) CABI compendium: invasive species Wallingford, UK: Centre for Agriculture and Biosciences International. Canna indica (Canna lilly). https://www.cabi.org/isc/datasheet/14575. Accessed 29 Mar 2023
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19(1):11–15
El-Tony FEIZH (2021) Phylogenetic relationship among different varieties and species of canna using molecular markers. Sci J Flowers Ornam Plants 8(1):165–180. https://doi.org/10.21608/SJFOP.2021.163280
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Gupta A, Maurya R, Roy RK, Sawant SV, Yadav HK (2013) AFLP based genetic relationship and population structure analysis of Canna-An ornamental plant. Sci Hortic 154:1–7. https://doi.org/10.1016/j.scienta.2013.02.005
Heywood VH (1993) Flowering plants of the World. Oxford University Press, New York, pp 296–299
Hermann M, Uptmoor R, Freire I, Montalvo JL (1997) Crop growth and starch productivity of edible Canna. CIP, Lima, Peru, pp 295–301
Imai K (2008) Edible Canna: a prospective plant resource from South America. J J Plant Sci 2(2):45–63
Jaccard P (1912) The distribution of the flora in the alpine zone. New Phytol 11(2):37–50. https://doi.org/10.1111/j.1469-8137.1912.tb05611.x
Khoshoo TN, Mukherjee I (1970a) Genetic–evolutionary studies on cultivated Cannas. III. Variation in phenotypes. Proc Indian Natl Sci 36B:254–270
Khoshoo TN, Mukherjee I (1970b) Genetic–evolutionary studies on cultivated Cannas. VI. Origin and evolution of ornamental taxa. Theor Appl Genet 40(5):204–212. https://doi.org/10.1007/BF00285243
Kranzlin FWL (1912) Das Pflanzenreich: IV. 47 Cannaceae. Leipzig: Verlag von Wilhelm Engelmann
Mass-van de Kamer H, Maas PJM (2008) The Cannaceae of the world. Blumea 53(2):247–318. https://doi.org/10.3767/000651908X607945
Mishra T, Goyal AK, Sen A (2018) Molecular profiling of 20 different accessions of Canna using RAPD and ISSR primers. NeBIO 9(2):180–187
Patra B, Acharya L, Mukherjee AK, Panda MK, Panda PC (2008) Molecular characterization of ten cultivars of Canna lilies (Canna Linn) using PCR based molecular markers (RAPDs and ISSRs). Int J Integr Biol 2(2):129–137
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research-an update. Bioinform 28(19):2537–2539. https://doi.org/10.1093/bioinformatics/bts460
Prevost A, Wilkinson MJ (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genetic 98:107–112
Prince LM (2010) Phylogenetic relationships and species delimitation in Canna (Cannaceae). Diversity, phylogeny, and evolution in the monocotyledons. Aarhus University Press, Denmark, pp 307–331
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959. https://doi.org/10.1093/genetics/155.2.945
Puncha-arnon S, Puttanlek C, Rungsardthong V, Pathipanawat W, Uttapap D (2007) Changes in physicochemical properties and morphology of Canna starches during rhizomal development. Carbohydr Polym 70(2):206–212. https://doi.org/10.1016/j.carbpol.2007.03.020
Rogers GK (1984) The zingiberales (Cannaceae, Marantaceae, and Zingiberaceae) in the Southeastern United States. J Arnold Arbor 65:29–39
Rohlf FJ (2002) NTSYS pc: numerical taxonomy system, Version 21. Exeter Publishing, Setauket, NY
Roldan-Ruiz I, Dendauw J, Vanbockstaele E, Depicker A, De Loose M (2000) AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol Breed 6:125–134
Sari N, Purnomo, Daryono BS, Suryadiantina, Setyowati M (2016) Variation and intraspecies classification of edible canna (Canna indica l.) based on morphological characters. Towards the sustainable use of biodiversity in a changing environment: From basic to applied research AIP Conf. Proc. 44. https://doi.org/10.1063/1.4953515
Sasaerila YH, Sakinah S, Noriko N, Wijihastuti RS (2021) Effects of light environments on leaf traits and phenotypic plasticity of Canna indica. Biosaintifika 13(2):169–177. https://doi.org/10.15294/biosaintifika.v13i2.305
Shete S, Tiwari H, Elston RC (2000) On estimating the heterozygosity and polymorphism information content value. Theor Popul Biol 57(3):265–271. https://doi.org/10.1006/tpbi.2000.1452
Tanaka N (1998) Economic botanical notes on edible Canna (Cannaceae) in South Vietnam. J Japanese Bot 73(6):319–324
Tanaka N (2001) Taxonomic revision of the family Cannaceae in the New World and Asia. Makinoa 1:1–74
Thitipraphunkul K, Uttapap D, Piyachomkwan K, Takeda Y (2003) A comparative study of edible canna (Canna edulis) starch from different cultivars. Part I: chemical composition and physicochemical properties. Carbohydr Polym 53(3):3–324. https://doi.org/10.1016/S0144-86(03)00081-X
Vaiman D, Mercier D, Moazai G (1994) A set of 99 cattle microsatellite, characterization, synteny mapping and polymorphism. Mamm Genome 5:288–297. https://doi.org/10.1007/BF00389543
Varshney RK, Chabane K, Hendre PS, Aggarwal RK, Graner A (2007) Comparative assessment of EST-SSR, EST-SNP and AFLP markers for evaluation of genetic diversity and conservation of genetic resources using wild, cultivated and elite barleys. Plant Sci 173:638–649
Zhu Q, Caib L, Lia H, Zhanga Y, Sua W, Zhou Q (2020) The complete chloroplast genome sequence of the Canna edulis Ker Gawl. (Cannaceae). Mitochondrial DNA Part B 5:2427–2428. https://doi.org/10.1080/23802359.2020.75512
Author Information
Plant Resources Center, Vietnam Academy of Agricultural Sciences, Hanoi, Vietnam