Genetic diversity in linseed (Linum usitatissimum) cultivars based on RAPD analysis
*Article not assigned to an issue yet
Research Articles | Published: 06 August, 2025
First Page: 0
Last Page: 0
Views: 79
Keywords: Linseed (Linum usitatissimum L.) (flax), Genetic diversity, RAPD, F1 u0026 F2 crosses and bud fly
Abstract
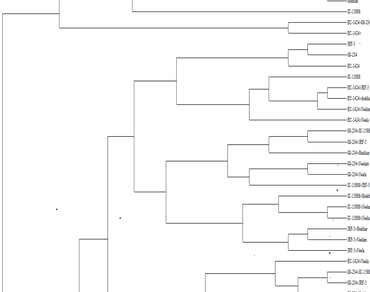
Identification of RAPD (Random Amplified Polymorphic DNA) marker for bud fly resistant gene. In the present study, 25 RAPD primers were tested to distinguish the parental lines, F1 and F2 lines. However, only 15 RAPD gave scorable and reproducible band. All these markers were found to be reproducible in three repeated RAPD. Data from these scorable primers were converted in to 0–1 data, based on the presence or absence of bands, these 0–1 data were used to calculate, jaccard’s similarity coefficient, upon which UPGMA cluster analysis were performed. The number of bands generated per primer ranged from 3 to 8 with a mean of 5.67 bands per primer. Out of 81 bands generated, 53 (65.43%) were polymorphic and 20 (24.69%) were monomorphic. The similarity coefficient of pair-wise comparisons among genotypes ranged from 0.48 to 1.00. The dendogram, generated from similarity matrix, was based on the principle of UPGMA. However, the dendogram showed only 30 genotypes, it may have happened the tie of some similar values. Based on the Jaccard similarity coefficient, four groups were obtained. The genetic similarity value was highest between Shekhar ×Neela and Neelam×Neela (1.00), whereas Neelam×IC15888 ×JRF, Neelam ×IC15888×Shekhar and Neela ×EC1424×JRF-5 showed the least similarity (0.294). The highest variability was in Neelam, while Neela was the least variable. The similarity coefficient in group I ranged from 0.647 to 0.970. By contrast, the similarity coefficient of group II ranged from 0.588 to 0.970. Group III showed a similarity coefficient range of 0.705–0.970. Group IV showed a similarity coefficient range of 0.911–0.970.
References
Abou El-Nasar THS, Hassanein MS, Ottai MES, Al-Kordy MA (2014) Genetic diversity among five Romanian linseed varieties under Egyptian conditions. Middle East J Appl Sci 4:114–121
Anonymous (2013) Production of Crops: Linseed: Area Harvested and Production (tonnes). Available on line http://faostat3.fao.org/home/index.html
Anonymous Ministry of Agriculture, Government of India; c2021-22
Bardakci F (2001) Random amplified polymorphic DNA (RAPD) markers. Turk J Biol 25:185–196
Betal S, Chowdhury PR, Kundu S, Raychaudhuri SS (2004) Estimation of genetic variability of Vigna radiata cultivars by RAPD analysis. Biol Plant 48:205–209
Chattopadhyay K, Bhattacharya S, Mandal N, Sarkar HK (2008) PCR-based characterization of Mungbean (Vigna radiata) genotypes from Indian Subcontinent at intra- and inter-specific level. J Plant Bio Biot 2:801–811
Dweikat I, Ohm H, Mackenzie S, Patterson F, Cambson S, Ratcliffe R (1994) Association of a DNA marker with Hessian fly resistance gene H9 in wheat. Teor Appl Genet 89:964–968
Dweikat I, Ohm H, Patterson F, Cambson S (1997) Identification of DNA markers for 11 Hessian fly resistance genes in wheat. Teor Appl Genet 94:419–423
Fu YBA, Diederichsen A, Richards KW, Richards, Peterson G (2001) Genetic diversity within a range of cultivars and landraces of flax (Linum usitatissimum) as revealed by RAPDs. Genet Res Crop Evaluation 49(2):167–174
Fu YB, Rowland GG, Duguid SD, Richards KW (2003) RAPD analysis of 54 North American flax cultivars. Crop Sci 43:1084–1091
Fu YB, Gregory P, Diederichsen A, Richards KW (2002a) RAPD analysis of genetic relationships of seven flax species in the genus Linum L. Genetic resources and crop evolution. 49:253–259
Fu YB (2005) Geographic patterns of RAPD variation in cultivated flax. Crop Sci 45:1084–1091
Husain K, Bhatee YN, Srivastava RL, Chandra R, Malik YP, Dubey SD (2009) Crop weed competition in linseed (Linum usitatissimum L). Journal of Oilseeds Research, 26(spl. issue): 530–531
Ijaz A (2013) Anum s hahbaz; Ihsan ullah; Sadia ali; Tayyaba shaheen; Mahmood-ur-Rehman; Usman Ijaz and Sami ullah. (2013). Molecualar characterization of linseed germplasm using RAPD DNA fingerprinting markers. American-Eurasian J Agric Environ Sci 13(9):1266–1274
Lakhanpaul S, Chadha S, Bhat KV (2000) Random amplified polymorphic DNA (RAPD) analysis in Indian mung bean (Vigna radiata L.) cultivars. Genetica 109:227–234
Malik YP (1999) Varietal preference and economic injury level of bud fly (Dasyneura Lini Barnes) in linseed. J Oilseeds Res 16(1):97–100
Mani VS, Gautam KC, Chakraborty TK (1968) Losses in crop yield in India due to weed growth. PAN (c), 14: 142–158
Nair S, Bentur JS, Rao UP, Mohan M (1995) DNA markers tightly linked to a gall midge resistance gene (Gm2) are potentially useful for marker aided selection in rice. Theor Appl Genet 91:68–73
Nair S, Kumar A, Srivastava MN, Mohan M (1996) PCR-Based DNA markers linked to a gall midge resistance gene Gm4t, has potential for marker-aided selection in rice. Theor Appl Genet 92:660–665
Pali V, Mehta N (2016) Genetic diversity assessment of flax (Linum usitatissimum L.) germplasm using molecular and morphological markers. Electron J Plant Breed 7(4):986–995
Patnaik HP, Lenka D (2000) Damage potential of Helicoverpa armigera hubner in linseed. Indian J Agric Sci 70(3):197–198
Prabhakar KS, Akram M, Srinivas RL (2009) Genetic diversity in linseed (Linum usitatissimum L) cultivars based on RAPD analysis. Indian J Agric Sci 79:1046–1049
Ragupathy R, Rajkumar R, Cloutier S (2011) Physical mapping and BAC-end sequence analysis provide initial insights into the flax (Linum usitatissimum L.) genome. BMC Genomics 12:217
Reddy MP, Arsul BT, Shaik NR, Maheshwari JJ (2013) Estimation of heterosis for some traits in linseed (Linum usitatissimum L). ISOR J Agric Veterinary Sci (ISOR-JAVS) 2(5):11–17
Roche P, Alston FH, Maliepaard C, Evans KM, Vrielink R, Dunemann F, Markussen T, Tartarini S, Brown LM, Ryder C, King GJ (1997) RFLP and RAPD markers linked to the Rosy leaf curling aphid resistance gene (Sd1) in Apple. Theor Apple Genet 94:528–533
Saghai Marrof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphism in barley: Mendelian inheritance, chromosomal location and population dynamic. Proceedings of National Academy of Science USA, 81: 8014–8018
Saharan GS, Saharan MS (1999) Inheritance of resistance in linseed to powdery mildew (Odiuum lini) disease. Indian Phytopathol 52(1):86–87
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: a laboratory manual, 2nd edition. Cold Spring Harbor Laboratory Press, Plainview, New York, USA
Shrivastava N, Katiyar OP, Das SB, Shrivastava S (1994) Studies on evaluation of some germplasm line of linseed against bud fly (Dasyneura Lini Barnes). Geobios 21(2):77–81
Shrivastava RL, Singh J, Husain K, Malik YP, Dubey SD, Rai J, Bajpai M (1997) Linseed. In: Efficient Mamagement Of Drylalnd Crop in India-Oilseeds (Eds. Singh, R.P., Ready P.S. and Kiresur, V.), Publ. Indian Socity of Oilseeds Research, DOR, Hyderabad,pp 228–256
Singh PK, Mohd A, Srivastava RL (2009) Genetic diversity in linseed (Linum usitatissimum) cultivars based on RAPD analysis. Indian J Agric Sci 79(12):1046–1049
Williams JKG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Author Information
Meerut Institute of Technology, Meerut, Meerut, India