In silico molecular docking analysis and ADME prediction of cow urine derived 1-heneicosanol against some bacterial proteins
Short Communications | Published: 07 February, 2023
First Page: 426
Last Page: 432
Views: 3425
Keywords: Molecular docking, ADME, 1-Heneicosanol
Abstract
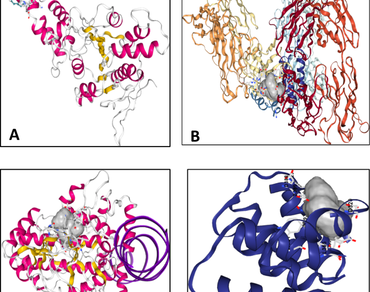
The continuously increasing drug resistance necessitate the search for new antibacterial drugs by exploiting natural resources. The lack of awareness and indiscriminate use of antibiotics are one of the major causes of the development of drug resistance. The present study was undertaken to evaluate the potential of a bioactive compound, 1-heneicosanol against selected pathogenic bacterial protein. In-silico molecular docking of 1-heneicosanol was carried out against four bacterial proteins viz., T3SS ATPases, SpA, PrfA and PelB. The results revealed considerable binding affinity to target proteins. Docking score of 1-heneicosanol against S. aureus was − 4.4 kcal/mol and − 4.3 kcal/mol, − 4.2 kcal/mol and − 3.9 kcal/mol against L. monocytogenes, P. aeruginosa and E. coli, respectively. Further, 1-heneicosanol was also investigated for ADME and drug likeness properties. The compound showed acceptable ADME profile and also followed Lipinski rule of five. The outcome of this study suggests that 1-heneicosanol could be considered as a potential antibacterial agent of animal origin which can reduce the risk of drug resistance.
References
Ahmad S, Zahiruddin S, Parvee B, Basist P, Parveen A, Parveen R et al (2021) Indian medicinal plants and formulations and their potential against COVID-19—preclinical and clinical research. Front Pharmacol 11:2470
Allam AE, Amen Y, Ashour A, Assaf HK, Hassan HA, Abdel-Rahman IM et al (2021) In silico study of natural compounds from sesame against COVID-19 by targeting M pro. PL pro and RdRp. RSC Adv 11(36):22398–22408
Dramsi S, Lebrun M, Cossart P (1996) Molecular and genetic determinants involved in invasion of mammalian cells by Listeria monocytogenes. Curr Top Microbiol Immunol 209:61–77
Duncan MC, Linington RG, Auerbuch V (2012) Chemical inhibitors of the type three secretion system: disarming bacterial pathogens. Antimicrob Agents Chemother 56(11):5433–5441
Forsgren A, Sjöquist J (1996) “Protein a” from S. aureus: I. Pseudo-immune reaction with human γ-globulin. J Immunol 97(6):822–827
Ge H, Wang Y, Zhao X (2022) Research on the drug resistance mechanism of foodborne pathogens. Microb Pathog 162:105306
Ghadaksaz A, Fooladi AAI, Hosseini HM, Amin M (2015) The prevalence of some Pseudomonas virulence genes related to biofilm formation and alginate production among clinical isolates. J Appl Biomed 13(1):61–68
Gupta KK, Aneja KR, Rana D (2016) Current status of cow dung as a bioresource for sustainable development. Bioresour Bioprocess 3(1):1–11
Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K et al (2001) Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res 8(1):11–22
Joshi R, Reeta KH, Sharma SK, Tripathi M, Gupta YK (2015) Panchagavya Ghrita, an Ayurvedic formulation attenuates seizures, cognitive impairment and oxidative stress in pentylenetetrazole induced seizures in rats. Indian J Exp Biol 53:446–451
Kaplan JL, Shi HN, Walker WA (2011) The role of microbes in developmental immunologic programming. Pediatr Res 69(6):465–472
Khan MF, Kader FB, Arman M, Ahmed S, Lyzu C, Sakib SA, Capasso R (2020) Pharmacological insights and prediction of lead bioactive isolates of Dita bark through experimental and computer-aided mechanism. Biomed Pharmacother 131:110774
Klevens RM, Edwards JR, Gaynes RP (2008) The impact of antimicrobial-resistant, health care-associated infections on mortality in the United States. Clin Infect Dis 47(7):927–930
Lipinski CA (2004) Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 1(4):337–341
Luyt CE, Bréchot N, Trouillet JL, Chastre J (2014) Antibiotic stewardship in the intensive care unit. Crit Care 18(5):1–12
Milohanic E, Glaser P, Coppée JY, Frangeul L, Vega Y, Jose A et al (2003) Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol Microbiol 47(6):1613–1625
Moo CL, Yang SK, Yusoff K, Ajat M, Thomas W, Abushelaibi A (2020) Mechanisms of antimicrobial resistance (AMR) and alternative approaches to overcome AMR. Curr Drug Discov Technol 17(4):430–447
Nadendla RR (2004) Molecular modeling: a powerful tool for drug design and molecular docking. Resonance 9(5):51–60
Nautiyal V, Dubey RC (2021) FT-IR and GC–MS analyses of potential bioactive compounds of cow urine and its antibacterial activity. Saudi J Biol Sci 28(4):2432–2437
Pamer EG (2007) Immune responses to commensal and environmental microbes. Nat Immunol 8(11):1173–1178
Pradhan S, Dubey RC (2022) Deciphering antimicrobial, phytochemical, GC–MS and pharmacokinetic properties of Camellia sinensis from high-altitude region. Vegetos 35:895–902
Pradhan S, Dubey RC (2021) Molecular docking of a bioactive compound of C. sinensis n-heptadecanol-1 with opportunistic fungi. Curr Res Green Sustain Chem 4:100208
Rhetso T, Shubharani R, Roopa MS, Sivaram V (2020) Chemical constituents, antioxidant, and antimicrobial activity of Allium chinense G. Don. Future J Pharm Sci 6(1):1–9
Slater SL, Sågfors AM, Pollard DJ, Ruano-Gallego D, Frankel G (2018) The type III secretion system of pathogenic Escherichia coli. Curr Top Microbiol Immunol 416:51–72
Suk KH, Gopinath SC, Anbu P, Lakshmipriya T (2018) Cellulose nanoparticles encapsulated cow urine for effective inhibition of pathogens. Powder Technol 328:140–147
Author Information
Department of Botany and Microbiology, Gurukula Kangri (Deemed to be University), Haridwar, India