In-silico prediction of microRNA targets and finding genes suggesting significant involvement in the development of Glycine max seed
Nivedita Yadav, Kavita Goswami, Budhayash Gautam, Pramod Kumar Yadav
Research Articles | Published: 06 November, 2019
First Page: 450
Last Page: 463
Views: 3924
Keywords: miRNAs, Degradome analysis, miRNA target prediction, GO analysis, Expression pattern
Abstract
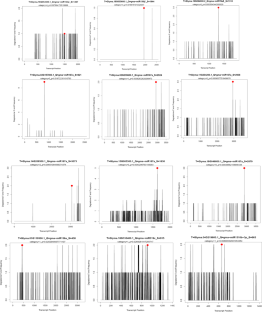
Glycine max is a worldwide leading economic crop and its seeds are deepening with proteins and oils which supply food and sustenance to all being. Various amounts of alimentary constituents are racked up in the G. max seed in the period of its ontogenesis. Thus, grasping the regulation of biological functions during seed enlargement belong to the basics for crop enhancement. The gene regulatory characteristics of miRNAs in G. max attracted us to focus on its target gene prediction, gene ontology (GO) analysis and expression pattern to their miRNA target genes, which suggest significant involvement in the development of G. max seed. Seven miRNAs have been found from the differential gene expression analysis of development stage 0–4 mm vs. 12–16 mm of G. max seed on the statistical parameter of p value ≤ 0.05 by computational-based microarray data analysis for miRNA target gene prediction. The miRNA target prediction analysis showed total 23 genes that were cleaved from 6 miRNAs, and computationally validated by identifying t-plots of miRNA targets using CleaveLand tool. GO results confirmed that the differentially expressed target genes could be classified into 20 molecular function categories, 73 biological process categories, and 10 cell components categories. On the basis of GO results, two genes were found to be significantly involved in the developmental process of G.max seed. The first miRNA target gene Glyma.01g119500 was predicted to annotate for embryo development ending in seed dormancy, seed dormancy, seed maturation, and seed germination. The second miRNA target gene Glyma.15g005300 was found to be involved in the regulation of seed germination. The Soybean eFP browser analysis suggests that the gene Glyma.01g119500 and Glyma.15g005300 reaches its maximum expression level of 35.88 and 26.6 respectively in the Soybean data source. The present study provides an avenue to explore more genomic and proteomic information about G. max seed developmental stage-specific miRNA target genes.
References
- Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18(10):758–762. https://doi.org/10.1016/j.cub.2008.04.042
- Addo-Quaye C, Miller W, Axtell MJ (2009) CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25:130–131. https://doi.org/10.1093/bioinformatics/btn604
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP et al (2000) Gene Ontology: tool for the unification of biology. Nature Genet 25:25–29. https://doi.org/10.1038/75556
- Blein T, Patrick P (2016) MicroRNAs (miRNAs) and Plant Development. Wiley online library. https://doi.org/10.1002/9780470015902.a0020106.pub2
- Fahlgren N, Howell MD, Kasschau KD et al (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS One 2(2):e219. https://doi.org/10.1371/journal.pone.0000219
- Fu C, Sunkar R, Zhou C, Shen H et al (2012) Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol J 10:443–452. https://doi.org/10.1111/j.1467-7652.2011.00677.x
- Gai YP, Li YQ et al (2014) Analysis of phytoplasma-responsive sRNAs provide insight into the pathogenic mechanisms of mulberry yellow dwarf disease. Sci Rep 4:5378. https://doi.org/10.1038/srep05378
- Gaudet P, Dessimoz C (2017) Gene Ontology: Pitfalls, Biases, and Remedies. In: Dessimoz C, Skunca N (eds) The Gene Ontology Handbook. Methods Mol Biol, vol 1446. Humana Press, NY
- German MA, Pillay M, Jeong DH et al (2008) Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol 26:941–946. https://doi.org/10.1038/nbt1417
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34:D140–D144. https://doi.org/10.1093/nar/gkj112
- Guo C, Xu Y, Shi M et al (2017) Repression of miR156 by miR159 regulates the timing of the juvenile-to-adult transition in Arabidopsis. Plant Cell 29(6):1293–1304. https://doi.org/10.1105/tpc.16.00975
- Huang SQ, Xiang AL, Che LL et al (2010) A set of miRNAs from Brassica napus in response to sulphate deficiency and cadmium stress. Plant Biotechnol J 8(8):887–899. https://doi.org/10.1111/j.1467-7652.2010.00517.x
- Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14(6):787–799. https://doi.org/10.1016/j.molcel.2004.05.027
- Libault M, Farmer A, Joshi T et al (2010) An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 63:86–99. https://doi.org/10.1111/j.1365-313X.2010.04222.x
- Millar AA, Waterhouse PM (2005) Plant and animal microRNAs: similarities and differences. Funct Integr Genomics 5:129–135. https://doi.org/10.1007/s10142-005-0145-2
- Nakano M, Nobuta K et al (2006) Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res 34:D731–D735. https://doi.org/10.1093/nar/gkj077
- Schmutz J, Cannon SB, Schlueter J et al (2010) Genome sequence of the palaeopolyploid soybean. Nature 463(7278):178–183. https://doi.org/10.1038/nature08670
- Song QX, Liu YF, Hu XY et al (2011) Identification of miRNAs and their target genes in developing soybean seeds by deep sequencing. BMC Plant Biol 11:5. https://doi.org/10.1186/1471-2229-11-5
- Tripathi RK, Goel R, Kumari S, Dahuja A (2017) Genomic organization, phylogenetic comparison, and expression profiles of the SPL family genes and their regulation in soybean. Dev Genes Evol 227:101. https://doi.org/10.1007/s00427-017-0574-7
- Turner M, Yu O, Subramanian S (2012) Genome organization and characteristics of soybean microRNAs. BMC Genomics 13:169. https://doi.org/10.1186/1471-2164-13-169
- Wang Y, Wang Z, Amyot L, Tian L, Xu Z, Gruber MY, Hannoufa A (2014) Ectopic expression of miR156 represses nodulation and causes morphological and developmental changes in Lotus japonicus. Mol Genet Genomics 290:471–484. https://doi.org/10.1007/s00438-014-0931-4
- Wang Y, Li K, Chen L, Zou Y et al (2015) MicroRNA167-directed regulation of the auxin response factors GmARF8a and GmARF8b is required for soybean nodulation and lateral root development. Plant Physiol 168(3):984–999. https://doi.org/10.1104/pp.15.00265
- Wang Y, Lan Q, Zhao X, Xu W, Li F, Wang Q, Chen R (2016) Comparative profiling of microRNA expression in soybean seeds from genetically modified plants and their near-isogenic parental lines. PLoS One 11(5):e0155896. https://doi.org/10.1371/journal.pone.0155896
- Winter D, Vinegar B et al (2007) An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2(8):e718. https://doi.org/10.1371/journal.pone.0000718
- Wu G, Park MY, Conway SR, Wang JW et al (2009) The sequential action of miR156 and miR172 regulates developmental timing in arabidopsis. Cell 138:750–759. https://doi.org/10.1016/j.cell.2009.06.031
- Nivedita Yadav, Yadav PK, Gautam B (2015) Gene expression profiling of transcription factors of Arabidopsis thaliana using microarray data analysis. IJARCSSE 5(4):783–793
- Yadav N, Gautam B, Yadav PK (2019) Computational analysis of microarray-based gene expression profiling and unveiling the functional traits in the developmental phases of Glycine max seed. Vegetos. https://doi.org/10.1007/s42535-019-00008-5
- Yan Z, Hossain MS, Wang J, Valdes-Lopez O, Liang Y, Libault M, Qiu L, Stacey G (2013) miR172 regulates soybean nodulation. Mol Plant Microbe Interact 26:1371–1377. https://doi.org/10.1094/MPMI-04-13-0111-R
- Yin XC, Wang J, Cheng H, Wang XL, Yu DY (2013) Detection and evolutionary analysis of soybean miRNAs responsive to soybean mosaic virus. Planta 237:1213–1225. https://doi.org/10.1007/s00425-012-1835-3
Author Information
Jacob Institute of Biotechnology and Bioengineering, Department of Computational Biology and Bioinformatics, Sam Higginbottom University of Agriculture, Technology and Sciences, Prayagraj, India