Integrated breeding, genomics, and epigenetic approaches enhance aflatoxin resistance in peanut (Arachis hypogaea L.) under pathogen and climate stress
*Article not assigned to an issue yet
Review Articles | Published: 10 November, 2025
First Page: 0
Last Page: 0
Views: 164
Keywords: n A. flavusn , Aflatoxin resistance, Epigenetics, Genomics, Peanut
Abstract
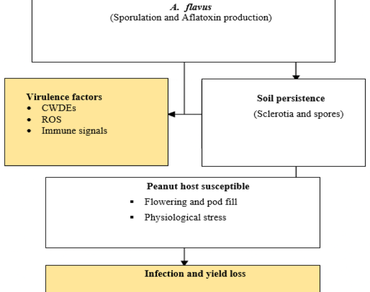
Aflatoxin contamination is a major constraint in global peanut (Arachis hypogaea L.) production, posing significant public health risks and trade restrictions. The toxin, primarily produced by A. flavus and A. parasiticus, is favored by specific environmental and physiological conditions, especially in drought-affected, tropical production systems. Recent advances in plant science reveal that an integrative strategy combining breeding, genomics, and epigenetics offers promising avenues for enhancing aflatoxin resistance. This review synthesizes current knowledge regarding the biological context pathogen behavior, climate impacts, and host plant interactions alongside modern resistance strategies. Under pathogen stress, A. flavus exploits weakened plant defenses and kernel microenvironments to produce aflatoxins. Climate-induced stress, especially drought and heat during pod filling, exacerbates fungal invasion and toxin accumulation. Traditional breeding has identified partial resistance sources, while genomic studies have pinpointed QTLs and candidate resistance genes. Epigenetic mechanisms, including DNA methylation and histone modifications, are emerging as novel regulatory systems that modulate gene expression in response to aflatoxin exposure. Strategically, marker-assisted selection, genome-wide association studies, CRISPR-based editing, and epigenome-wide profiling now enable more precise and durable resistance breeding. This review concludes that combining these tools under integrated frameworks holds the key to developing aflatoxin-resilient peanut cultivars adapted to increasingly variable climates and pathogen pressures.
References
Ahmad A, Jamil A, Munawar N (2023) Gmos or non-gmos? The CRISPR conundrum. Front Plant Sci 14:1232938. https://doi.org/10.3389/fpls.2023.1232938
Alam T (2025) Advances in tissue culture-free genetic engineering and genome editing of peanut. Mol Biotechnol. https://doi.org/10.1007/s12033-025-01476-8
Ali ML, Sanchez PL, Yu S-B, Lorieux M, Eizenga GC (2010) Chromosome segment substitution lines: a powerful tool for the introgression of valuable genes from Oryza wild species into cultivated rice (O. sativa). Rice 3:218–234. https://doi.org/10.1007/s12284-010-9058-3
Baldini A, Battaglia F, Perrella G (2025) The generation of novel epialleles in plants: the prospective behind re-shaping the epigenome. Front Plant Sci. https://doi.org/10.3389/fpls.2025.1544744
Bereziartua A, Huss A, Kers JG, Smit LAM, Vermeulen R, Figueiredo DM (2025) Pre-harvest aflatoxin contamination in crops and climate change factors: a European overview. Toxins. https://doi.org/10.3390/toxins17070344
Bhat RS, Rockey J, Shirasawa K, Tilak IS, Brijesh Patil MP, Reddy Lachagari VB (2020) DNA methylation and expression analyses reveal epialleles for the foliar disease resistance genes in peanut (Arachis hypogaea L.). BMC Res Notes 13:20. https://doi.org/10.1186/s13104-020-4883-y
Bressano M, Massa AN, Arias RS, de Blas F, Oddino C, Faustinelli PC, Soave S, Soave JH, Perez MA, Sobolev VS (2019) Introgression of peanut smut resistance from landraces to elite peanut cultivars (Arachis hypogaea L.). PLoS ONE 14:e0211920
Bunny SM, Umar A, Bhatti HS, Honey SF (2024) Aflatoxin risk in the era of climatic change—a comprehensive review. CABI Agric Biosci 5:105. https://doi.org/10.1186/s43170-024-00305-3
Caceres I, Al Khoury A, El Khoury R, Lorber S, P. Oswald I, El Khoury A, Atoui A, Puel O, Bailly J-D (2020) Aflatoxin biosynthesis and genetic regulation: a review. Toxins 12:150
Can SN, Nunn A, Galanti D, Langenberger D, Becker C, Volmer K, Heer K, Opgenoorth L, Fernandez-Pozo N, Rensing SA (2021) The epidiverse plant epigenome-wide association studies (EWAS) pipeline. Epigenomes. https://doi.org/10.3390/epigenomes5020012
Cao S, Chen ZJ (2024) Transgenerational epigenetic inheritance during plant evolution and breeding. Trends Plant Sci 29:1203–1223. https://doi.org/10.1016/j.tplants.2024.04.007
Chang Y, Gelaye Y, Yao R, Yang P, Li J, Liu N, Huang L, Zhou X, Chen W, Yu B (2025) Identification and characterization of histone modification gene families and their expression patterns during pod and seed development in peanut. Int J Mol Sci 26:2591
Chen Z, Grover CE, Ge X, Shangguan X (2023) Editorial: omics technology in agriculture: molecular breeding for sustainable crop production. Front Plant Sci. https://doi.org/10.3389/fpls.2023.1258701
Chen X, Xu H, Shu X, Song C-X (2025) Mapping epigenetic modifications by sequencing technologies. Cell Death Differ 32:56–65. https://doi.org/10.1038/s41418-023-01213-1
Chopra R, Burow G, Farmer A, Mudge J, Simpson CE, Wilkins TA, Baring MR, Puppala N, Chamberlin KD, Burow MD (2015) Next-generation transcriptome sequencing, SNP discovery and validation in four market classes of peanut, Arachis hypogaea L. Mol Genet Genomics 290:1169–1180. https://doi.org/10.1007/s00438-014-0976-4
Craufurd P, Prasad P, Waliyar F, Taheri A (2006) Drought, pod yield, pre-harvest Aspergillus infection and aflatoxin contamination on peanut in Niger. Field Crops Res 98:20–29
Dai Y, Huang K, Zhang B, Zhu L, Xu W (2017) Aflatoxin B1-induced epigenetic alterations: an overview. Food Chem Toxicol 109:683–689. https://doi.org/10.1016/j.fct.2017.06.034
de Vries RP, Visser J (2001) Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microbiol Mol Biol Rev 65:497–522. https://doi.org/10.1128/mmbr.65.4.497-522.2001
Ehrlich KC (2014) Non-aflatoxigenic Aspergillus flavus to prevent aflatoxin contamination in crops: advantages and limitations. Front Microbiol 5:50. https://doi.org/10.3389/fmicb.2014.00050
Faizal A, Nugroho S, Sembada AA, Theda Y, Komariyah T, Esyanti RR (2024) Genome editing in future crop protection: utilizing CRISPR/Cas9 to improve crop resistance against diseases, pests, and weeds. Discov Agric 2:104. https://doi.org/10.1007/s44279-024-00124-0
Fukushima A, Kanaya S, Nishida K (2014) Integrated network analysis and effective tools in plant systems biology. Front Plant Sci. https://doi.org/10.3389/fpls.2014.00598
Gangurde SS, Korani W, Bajaj P, Wang H, Fountain JC, Agarwal G, Pandey MK, Abbas HK, Chang P-K, Holbrook CC, Kemerait RC, Varshney RK, Dutta B, Clevenger JP, Guo B (2024) Aspergillus flavus pangenome (AflaPan) uncovers novel aflatoxin and secondary metabolite associated gene clusters. BMC Plant Biol 24:354. https://doi.org/10.1186/s12870-024-04950-8
George ME, Gaitor TT, Cluck DB, Henao-Martínez AF, Sells NR, Chastain DB (2025) The impact of climate change on the epidemiology of fungal infections: implications for diagnosis, treatment, and public health strategies. Ther Adv Infect Dis 12:20499361251313841. https://doi.org/10.1177/20499361251313841
Globus R, Qimron U (2018) A technological and regulatory outlook on CRISPR crop editing. J Cell Biochem 119:1291–1298. https://doi.org/10.1002/jcb.26303
Guchi E (2015) Aflatoxin contamination in groundnut (Arachis hypogaea L.) caused by Aspergillus species in Ethiopia. J Appl Environ Microbiol 3:11–19
Hasan N, Choudhary S, Naaz N, Sharma N, Laskar RA (2021) Recent advancements in molecular marker-assisted selection and applications in plant breeding programmes. J Genet Eng Biotechnol 19:128. https://doi.org/10.1186/s43141-021-00231-1
Hill RA, Blankenship PD, Cole RJ, Sanders TH (1983) Effects of soil moisture and temperature on preharvest invasion of peanuts by the Aspergillus flavus group and subsequent aflatoxin development. Appl Environ Microbiol 45:628–633. https://doi.org/10.1128/aem.45.2.628-633.1983
Horn BW (2005) Colonization of wounded peanut seeds by soil fungi: selectivity for species from Aspergillus section Flavi. Mycologia 97:202–217. https://doi.org/10.3852/mycologia.97.1.202
Huai D, Huang L, Xue X, Yu B, Ding Y, Jin G, Liu H, Pandey MK, Sudini HK, Luo H, Zhou X, Liu N, Chen W, Yan L, Chen Y, Wang X, Wang Q, Kang Y, Wang Z, Chen X, Jiang H, Lei Y, Liao B (2025) Identification of candidate genes associated with resistance to aflatoxin production in peanut through genetic mapping and transcriptome analysis. Theor Appl Genet 138:71. https://doi.org/10.1007/s00122-025-04822-1
Jaskiewicz M, Conrath U, Peterhänsel C (2011) Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep 12:50–55. https://doi.org/10.1038/embor.2010.186
Jiang Y, Luo H, Yu B, Ding Y, Kang Y, Huang L, Zhou X, Liu N, Chen W, Guo J, Huai D, Lei Y, Jiang H, Yan L, Liao B (2021) High-density genetic linkage map construction using whole-genome resequencing for mapping QTLs of resistance to Aspergillus flavus infection in peanut. Front Plant Sci. https://doi.org/10.3389/fpls.2021.745408
Kumar S, Mohapatra T (2021) Dynamics of DNA methylation and its functions in plant growth and development. Front Plant Sci. https://doi.org/10.3389/fpls.2021.596236
Lewis MH, Carbone I, Luis JM, Payne GA, Bowen KL, Hagan AK, Kemerait R, Heiniger R, Ojiambo PS (2019) Biocontrol strains differentially shift the genetic structure of indigenous soil populations of Aspergillus flavus. Front Microbiol. https://doi.org/10.3389/fmicb.2019.01738
Liu C, Van der Fels-Klerx HJ (2021) Quantitative modeling of climate change impacts on mycotoxins in cereals: a review. Toxins. https://doi.org/10.3390/toxins13040276
Liu J, Zhong X (2024) Epiallelic variation of non-coding RNA genes and their phenotypic consequences. Nat Commun 15:1375. https://doi.org/10.1038/s41467-024-45771-5
Liu L, Sonbol F-M, Huot B, Gu Y, Withers J, Mwimba M, Yao J, He SY, Dong X (2016) Salicylic acid receptors activate jasmonic acid signalling through a non-canonical pathway to promote effector-triggered immunity. Nat Commun 7:13099. https://doi.org/10.1038/ncomms13099
Mundt CC (2018) Pyramiding for resistance durability: theory and practice. Phytopathology 108:792–802
Nigam S, Waliyar F, Aruna R, Reddy S, Kumar PL, Craufurd PQ, Diallo A, Ntare B, Upadhyaya HD (2009) Breeding peanut for resistance to aflatoxin contamination at ICRISAT. Peanut Sci 36:42–49
Pasupuleti J, Nigam SN, Pandey MK, Nagesh P, Varshney RK (2013) Groundnut improvement: use of genetic and genomic tools. Front Plant Sci. https://doi.org/10.3389/fpls.2013.00023
Pothula S (2015) Genetic analysis of drought tolerance in groundnut (Arachis hypogaea. L), University of Agricultural Sciences, Dharwad.
Reddy DVR, Thirumala-Devi K (2003) Peanuts. In: Loebenstein G, Thottappilly G (eds) Virus and virus-like diseases of major crops in developing countries. Springer, Dordrecht, pp 397–423
Scott MF, Ladejobi O, Amer S, Bentley AR, Biernaskie J, Boden SA, Clark M, Dell’Acqua M, Dixon LE, Filippi CV, Fradgley N, Gardner KA, Mackay IJ, O’Sullivan D, Percival-Alwyn L, Roorkiwal M, Singh RK, Thudi M, Varshney RK, Venturini L, Whan A, Cockram J, Mott R (2020) Multi-parent populations in crops: a toolbox integrating genomics and genetic mapping with breeding. Heredity 125:396–416. https://doi.org/10.1038/s41437-020-0336-6
Seidel D, Wurster S, Jenks JD, Sati H, Gangneux JP, Egger M, Alastruey-Izquierdo A, Ford NP, Chowdhary A, Sprute R, Cornely O, Thompson GR 3rd, Hoenigl M, Kontoyiannis DP (2024) Impact of climate change and natural disasters on fungal infections. Lancet Microbe 5:e594–e605. https://doi.org/10.1016/s2666-5247(24)00039-9
Shah H, Hellegers P, Siderius C (2021) Climate risk to agriculture: a synthesis to define different types of critical moments. Clim Risk Manag 34:100378. https://doi.org/10.1016/j.crm.2021.100378
Shankar J, Tiwari S, Shishodia SK, Gangwar M, Hoda S, Thakur R, Vijayaraghavan P (2018) Molecular insights into development and virulence determinants of Aspergilli: a proteomic perspective. Front Cell Infect Microbiol 8:180. https://doi.org/10.3389/fcimb.2018.00180
Shi Z, Liu S, Noe J, Arelli P, Meksem K, Li Z (2015) Snp identification and marker assay development for high-throughput selection of soybean cyst nematode resistance. BMC Genomics 16:314. https://doi.org/10.1186/s12864-015-1531-3
Stadlmeier M, Hartl L, Mohler V (2018) Usefulness of a multiparent advanced generation intercross population with a greatly reduced mating design for genetic studies in winter wheat. Front Plant Sci. https://doi.org/10.3389/fpls.2018.01825
Sun D, Wei Y, Han C, Li X, Zhang Z, Wang S, Zhou Z, Gao J, Chen J, Wu J (2025) Genome-wide association study and RNA-seq elucidate the genetic mechanisms behind aphid (Rhopalosiphum maidis F.) resistance in maize. Plants. https://doi.org/10.3390/plants14111614
Suppa RW, Andres RJ, Dunne JC, Arram RF, Morgan TB, Chen H (2024) Autotetraploid induction of three A-genome wild peanut species, Arachis cardenasii, A. correntina, and A. diogoi. Genes 15:303
Ting WTE, Chang C-H, Szonyi B, Gizachew D (2020) Growth and aflatoxin B1, B2, G1, and G2 production by Aspergillus flavus and Aspergillus parasiticus on ground flax seeds (Linum usitatissimum). J Food Prot 83:975–983. https://doi.org/10.4315/JFP-19-539
Umar A, Bhatti HS, Honey SF (2023) A call for aflatoxin control in Asia. CABI Agric Biosci 4:27. https://doi.org/10.1186/s43170-023-00169-z
Varotto S, Tani E, Abraham E, Krugman T, Kapazoglou A, Melzer R, Radanović A, Miladinović D (2020) Epigenetics: possible applications in climate-smart crop breeding. J Exp Bot 71:5223–5236. https://doi.org/10.1093/jxb/eraa188
Varshney RK, Pandey MK, Janila P, Nigam SN, Sudini H, Gowda MV, Sriswathi M, Radhakrishnan T, Manohar SS, Nagesh P (2014) Marker-assisted introgression of a QTL region to improve rust resistance in three elite and popular varieties of peanut (Arachis hypogaea L.). Theor Appl Genet 127:1771–1781. https://doi.org/10.1007/s00122-014-2338-3
Vishwakarma K, Kumar N, Shandilya C, Mohapatra S, Bhayana S, Varma A (2020) Revisiting plant-microbe interactions and microbial consortia application for enhancing sustainable agriculture: a review. Front Microbiol. https://doi.org/10.3389/fmicb.2020.560406
Wang K, Yan PS, Ding QL, Wu QX, Wang ZB, Peng J (2013) Diversity of culturable root-associated/endophytic bacteria and their chitinolytic and aflatoxin inhibition activity of peanut plant in China. World J Microbiol Biotechnol 29:1–10. https://doi.org/10.1007/s11274-012-1135-x
Wang Y, Liu D, Yin H, Wang H, Cao C, Wang J, Zheng J, Liu J (2023) Transcriptomic and metabolomic analyses of the response of resistant peanut seeds to Aspergillus flavus infection. Toxins. https://doi.org/10.3390/toxins15070414
Williams SL, Toda M, Chiller T, Brunkard JM, Litvintseva AP (2024) Effects of climate change on fungal infections. PLoS Pathog 20:e1012219. https://doi.org/10.1371/journal.ppat.1012219
Wu F, Guclu H (2012) Aflatoxin regulations in a network of global maize trade. PLoS ONE 7:e45151. https://doi.org/10.1371/journal.pone.0045151
Wu F, Bhatnagar D, Bui-Klimke T, Carbone I, Hellmich R, Munkvold G, Paul P, Payne G, Takle E (2011) Climate change impacts on mycotoxin risks in US maize. World Mycotoxin J 4:79–93
Wu D, Yang C, Yao Y, Ma D, Lin H, Hao L, Xin W, Ye K, Sun M, Hu Y, Yang Y, Zhuang Z (2024) SntB triggers the antioxidant pathways to regulate development and aflatoxin biosynthesis in Aspergillus flavus. Elife. https://doi.org/10.7554/eLife.94743
Wu D, Zhao C, Korani W, Thompson EA, Wang H, Agarwal G, Fountain JC, Culbreath A, Holbrook CC, Wang X, Clevenger JP, Guo B (2025a) High-resolution genetic and physical mapping reveals a peanut spotted wilt disease resistance locus, PSWDR-1, to Tomato spotted wilt virus (TSWV), within a recombination cold-spot on chromosome A01. BMC Genomics 26:224. https://doi.org/10.1186/s12864-025-11366-7
Wu H, Yan H, Li B, Han Y, Liu N, Fan M, Liu Y, Lyu M, Du S, Wei A (2025b) GWAS and RNA-seq reveal novel loci and genes of low-nitrogen tolerance in cucumber (Cucumis sativus L.). Front Plant Sci. https://doi.org/10.3389/fpls.2025.1602360
Xu J, Pan Y, Peter RM, Chou PJ, Dave PD, Shanner A, Sarwar MS, Brunetti L, Simon JE, Kong AT (2025) Exploring the epigenetic and metabolic pathways for antioxidant and anti-inflammatory potentials of tart cherry juice concentrate. Curr Pharmacol Rep 11:43. https://doi.org/10.1007/s40495-025-00422-1
Yan L, Song W, Wang Z, Yu D, Sudini H, Kang Y, Lei Y, Huai D, Chen Y, Wang X, Wang Q, Liao B (2023) Dissection of the genetic basis of resistance to stem rot in cultivated peanuts (Arachis hypogaea L.) through genome-wide association study. Genes. https://doi.org/10.3390/genes14071447
Yu B, Huai D, Huang L, Kang Y, Ren X, Chen Y, Zhou X, Luo H, Liu N, Chen W, Lei Y, Pandey MK, Sudini H, Varshney RK, Liao B, Jiang H (2019) Identification of genomic regions and diagnostic markers for resistance to aflatoxin contamination in peanut (Arachis hypogaea L.). BMC Genet 20:32. https://doi.org/10.1186/s12863-019-0734-z
Zhang C, Xie W, Fu H, Chen Y, Chen H, Cai T, Yang Q, Zhuang Y, Zhong X, Chen K, Gao M, Liu F, Wan Y, Pandey MK, Varshney RK, Zhuang W (2022) Whole genome resequencing identifies candidate genes and allelic diagnostic markers for resistance to Ralstonia solanacearum infection in cultivated peanut (Arachis hypogaea L.). Front Plant Sci 13:1048168. https://doi.org/10.3389/fpls.2022.1048168
Author Information
Department of Horticulture, College of Agriculture and Natural Resources, Debre Markos University, Debre Markos, Ethiopia