Karyotype Stasis and Genetic Diversity in Amomum spp. from Tripura, North-East India
Research Article | Published: 11 January, 2019
First Page: 52
Last Page: 58
Views: 4564
Keywords: Amomum; Karyotype; Simple Sequence Repeat (SSR); Genetic diversity; Unweighted Pair Group Method with Arithmetic Mean (UPGMA); Tripura
Abstract
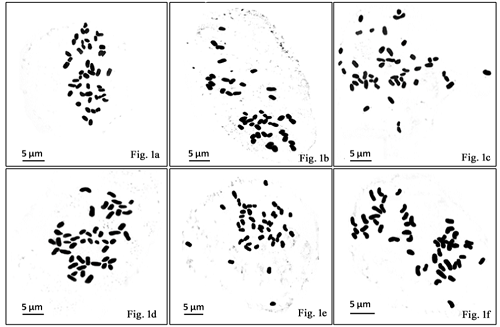
The genus Amomum of Zingiberaceae comprising a large number of aromatic plants, with its main centre of distribution in North Eastern India, is facing the threat of extinction due to anthropogenic disturbances. In this study, the somatic chromosome number of two wild populations with each of three species of Amomum viz. A. maximum, A. corynostachyum and A. aromaticum from the state of Tripura was determined by modifying aceto-orcein staining technique. Additionally, genetic diversity was assessed using Simple Sequence Repeat (SSR) molecular markers. The somatic chromosome count was 2n=48 with one pair of nearly metacentric chromosomes bearing secondary constriction noted in all the species even at the population level. A considerable homogeneity in their karyotype was observed both at interspecific and intraspecific level suggesting a karyotype stasis. In contrast, SSR analyses indicated a high degree of genetic polymorphism (75.00-100.00%). UPGMA dendrogram revealed two different clusters with inherent variability between the populations. Our findings suggest that the morphological distinction among the species, which could not be resolved at cytological level probably due to intrinsic characteristic of the karyotype itself, can be assessed through SSR fingerprinting.
References
- Basak S, Ramesh AM, Kesari V, Parida A, Mitra S and Rangan L. (2014). Genetic diversity relationship of Hedychium from Northeast India as dissected using PCA analysis and hierarchical clustering. Meta Gene 2: 459-468.
- Chakravorti AK (1952) Cytogenetical studies in Zingiberaceae. Proceeding of 39th Indian Science Congress part 3.
- Chen ZY, Chen SJ, Huang XX and Huang SF (1988) A report on chromosome numbers on Chinese Zingiberaceae. Guihaia 8:143-147.
- Das A, Kesari V, Satyanarayana VM, Parida A, Mitra S and Rangan L (2013) Genetic diversity in ecotypes of the scarce wild medicinal crop Zingiber moran revealed by ISSR and AFLP marker analysis and chromosome number assessment. Plant Biosystem 149 (1):111-120.
- Deb DB (1983) The flora of Tripura. New Delhi: Today and Tomorrow’s Printers and Publishers.
- Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26:297-302.
- Fernandez ME, Figueiras AM and Benito C (2002) The use of ISSR and RAPD markers for detecting DNA polymorphism, genotype identification and genetic diversity among barley cultivars with known origin. Theor Appl Genet 104: 845-851.
- Islam MA, Meister A, Schubert V, Kloppstech K and Esch E (2007) Genetic diversity and cytogenetic analysis in Curcuma zedoaria (Christm.) Roscoe. from Bangladesh. Genet Resour Crop Evol 54:149–156.
- Jain SK and Prakash V (1995) Zingiberaceae in India: phytogeography and endemism. Rheedea 5: 154-169.
- Jatoi SA, Kikuchi A, Yi SS, Naing KW, Yamanaka S and Junko A (2006) Use of SSR markers as RAPD markers for genetic diversity analysis in Zingiberaceae. Breed Sci 56:107–111.
- Kimura M and Crow JF (1964) The number of alleles that can be maintained in a finite population. Genetics 49:725-738.
- Kress WJ, Prince LM and Williams KJ (2002) The phylogeny and a new classification of the gingers (Zingiberaceae): evidence from molecular data. Am J Bot 89:1682-1696.
- Lamxay V and Newman MF (2012) A revision of Amomum (zingiberaceae) in cambodia, laos and vietnam. Edinb J Bot 69: 99–206.
- Lee SY, Fai WK, Zakaria M, Ibrahim H, Othman RY Gwag JG, Rao VR and Park YJ (2007) Characterization of polymorphic microsatellite markers, isolated from ginger (Zingiber officinale Rosc.). Mol Ecol Notes 7:1009–1011.
- Levan A, Fredga K and Sandbery AA (1964) Nomenclature for centromeric position on chromosomes. Heriditas 52:201-220.
- Lewontin RC (1972) Testing the theory of natural selection. Nature 236:181-182.
- Mandal B, Vijayanandraj S, Shilpi S, Pun KB, Singh V, Pant RP, Jain RK, Varadarasan S and Varma A (2012) Disease distribution and characterisation of a new macluravirus associated with chirke disease of large cardamom. Ann Appl Biol 160: 225–236.
- Mao AA, Hynniewta TM and Sanjappa M. (2009) Plant wealth of northeast India with reference to ethnobotany. Indian Journal of Traditional Knowledge 8:96-103.
- Mason AS (2015) SSR Genotyping. In: Batley J (ed) Plant Genotyping. Springer, 77-89.
- Myers N, Mittermier RA, Mittermier CG, Fonseca GABda and Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 40:853-858.
- Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci- USA 70:3321-3323.
- Pérez-Jiménez M, Besnard G, Dorado G and Hernandez P (2013) Varietal tracing of virgin olive oils based on plastid DNA variation profiling. PLoS One. 8.
- Phong DT, Hien VT, Thanh TT and Tang DV (2011) Comparison of RAPD and ISSR markers for assessment of genetic diversity among endangered rare Dalbergia oliveri (Fabaceae) genotypes in Vietnam. Genet Mol Res 10: 2382-2393.
- Phumichai C, Phumichai T and Wongkaew A (2015) Novel chloroplast microsatellite (cpSSR) markers for genetic diversity assessment of cultivated and wild Hevea rubber. Plant Mol Biol Report 33:1486–1498.
- Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S and Rafalski A (1999) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2: 225–238.
- Ramachandran K (1969) Chromosome numbers in zingiberaceae. Cytologia 34: 213-221.
- Rohlf FJ (2009) NTSYSpc: numerical taxonomy system. version 2.21c. Exeter Software: Setauket: New York.
- Roldan IR, Dendauw J, Van EB, Depicker A and Loose MD (2000) AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol Breed 6:125-134.
- Sabu M (2006) Zingiberaceae and costaceae of south India. Indian association for angiosperm taxonomy. India.
- Sharma AK and Bhattacharyya NK (1959) Cytology of several members of zingiberaceae. Cellule 59:297-346.
- Sharma AK and Sharma A (1980) Chromosome technique theory and practical. (3rd edition), Butterworth & Co (Publishers) Ltd. London.
- Siju S, Dhanya K, Syamkumar S, Sheeja TE, Sasikumar B, Bhat AI and Parthasarathy VA (2010) Development, characterization and utilization of genomic microsatellite markers in turmeric (Curcuma longa L.). Biochemical Systematics and Ecology 38: 641–646.
- Sneath PHA and Sokal RR (1973) Numerical taxonomy: the principles and practice of numerical classification.W. H. Freeman and company, San Francisco.
- Sokal RR and Michener CD (1958) A statistical method for evaluating systematic relationships. Sci Bull 38:1409-1438.
- Stebbins GL (1971) Chromosomal Evolution in Higher Plants. Addison- Wesley Publication Company, California.
- Thomas VP and Sabu M (2012) Two new species of Amomum (Zingiberaceae) from western ghats, India. Edinb J Bot 69:313–321.
- Venkatasubban KR (1946) A preliminary survey of chromosome numbers in scitamineae of Bentham and Hooker. Proceedings of Indian Academy of Science sec B23 p. 281-300.
- Yeh FC, Yang RC, Boyle TBJ, Ye ZH and Mao JX (1997) Popgene, the user friendly shareware for population genetic analysis.Version 1.32. Molecular Biology and Biotechnology Centre, University of Alberta.
- Zietkiewicz E, Rafalski A and Labuda D (1994) Genome fingerprinting by simple sequence repeat (SSR)-anchored polymerase chain reaction amplification. Genomics 20:176-183.
Author Information
Cytogenetics and Plant Biotechnology Laboratory, Department of Botany, Tripura University, Suryamaninagar-799022, Tripura, India,