Molecular characterization of cotton genotypes using PCR based molecular markers
*Article not assigned to an issue yet
Shinde C. S., Shitole L. S., Bhor T. J., Jadhav D. P., Nimbalkar R. D., Wagh R. S.
Research Articles | Published: 29 May, 2025
First Page: 0
Last Page: 0
Views: 970
Keywords: Diversity, SSR, Fiber quality, Markers, Similarity matrix, Correlation
Abstract
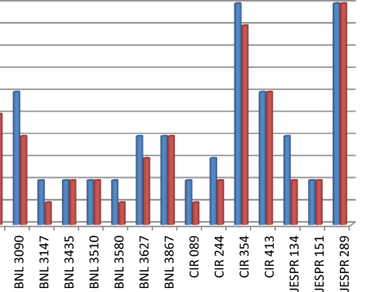
Total 25 cotton accessions and 25 SSR markers were selected to depict diversity and correlation between the genotypes for fiber quality traits. The existing study was conducted at Biotechnology Laboratory College of Agriculture, Pune and Central laboratory of ICAR-Directorate of Floricultural Research, Keshavnagar, Pune. Out of 25 SSR markers, 17 were amplified and all the markers were polymorphic. Total 71 loci amplified by 17 markers. Out of these, 59 were polymorphic and 12 were monomorphic. The polymorphic information content (PIC) value ranged from 0.18 to 0.82, maximum PIC recorded for JESPR-289 and minimum for CIR-089. The similarity matrix and dendrogram was constructed using Jaccard’s similarity coefficient with NTSYS 2.02i software. The value of coefficient of similarity varied from 0.46 to 0.97. The dendrogram revealed that two major clusters were formed among 25 cotton genotypes. Cluster I contain twenty one genotypes while Cluster II contain four genotypes. The correlation was calculated. Total of 7 markers were found to be correlated with fiber quality. Out of that seven markers, BNL-3090 and BNL-3867 were found to positively correlated with all three fiber quality parameters. The remaining five BNL-1059, BNL-3147, BNL-3435, BNL-3510 and CIR-413 were positively correlated with length and strength. BNL-3090 and BNL-3867 are only markers found to be correlated with fineness. These molecular markers can be effectively used in future cotton improvement program as well as integrated with DUS descriptor data supporting with DNA fingerprinting analysis.
References
Ali MA, Khan IA, Awan SI, Ali S, Niaz S (2008) Genetics of fiber quality traits in cotton. Aus J Crop Sci 2:10–17
Ambreen I, Sadia A, Usman IS, Tayyaba S (2013) Molecular characterization of cotton using simple sequence repeat (SSR) markers and application of genetic analysis. Int J Genet Mol Biol 5(4):49–53. https://doi.org/10.5897/IJGMB2013.0066
Anjani A, Padma V, Ramana JV, Satish Y (2020) Molecular characterization of cotton using simple sequence repeat (SSR) markers. Int J Chem Stud 8(4):3518–3522. https://doi.org/10.22271/chemi.2020.v8.i4ar.10197
Chavhan MT, Rathod DR, Taynath BS, Bhojane PN, Deshmukh SB, Gahukar SJ, Bhande AS, Mohanapure PA (2021) Molecular characterization of desi cotton hybrids using SSR markers for boll size improvement. Int J Curr Microbiol Appl Sci 10(01):113–123. https://doi.org/10.20546/ijcmas.2021.1001.013
Chen G, Du XM (2006) Genetic diversity of source germplasm of Upland cotton in China as determined by SSR marker analysis. Yi Chuan Xue Bao 33(8):733–745. https://doi.org/10.1016/S0379-4172(06)60106-6
Committee on Cotton Production and Consumption (COCPC) meeting held on 12th Nov 2021.
Doyle JJ, Doyle JL (1990) A rapid total DNA preparation procedure for fresh plant. Focus 12:13–15
Ezhilkumar S, Padmavathi S (2016) Identification of cotton (Gossypium spp.) genotypes by single sequence repeat (SSR) markers. Int J Adv Multidiscip 3(11):62–69. https://doi.org/10.22192/ijamr.2016.03.11.005
Ghuge SB, Mehetre SS, Chimote VP, Pawar BD, Naik RM (2018) Molecular characterization of cotton genotypes using SSR, ISSR and RAPD markers in relation to fiber quality traits. J Cotton Res Dev 32(1):1–12
Gillham EM, Thomas MB, Tije A, Matthews GA, Cluade LE, Rumeur CL, Brian HA (1995) Cotton prod. pros. for the next decade. World Bank Technical Paper Number-267.
Kotchoni SO, Gachomo EW (2009) A rapid and hazardous reagent free protocol for genomic DNA extraction suitable for genetic studies in plants. Mol Biol Rep 36:1633–1636. https://doi.org/10.1007/s11033-008-9362-9
Kumbhalkar HB, Gawande VL, Gahukar SJ, Waghmare VN, Mawle SR, Ingle KP (2018) Molecular profiling of cotton genotypes for fiber properties using diagnostic set of microsatellite (SSR) markers. Cur J Appl Sci 29(3):1–16. https://doi.org/10.9734/CJAST/2018/44497
Lacape JM, Llewellyn D, Jacobs J, Arioli T, Becker D, Calhoun S, Alghazi Y, Liu S, Palai O, Georges S (2010) Meta-analysis of cotton fiber quality QTLs. BMC Plant Biol 10:132
Liu D, Guo X, Lin Z, Nie Y, Zhang X (2005) Genetic diversity of Asian cotton (Gossypium arboreum L.) in China evaluated by microsatellite analysis. Genet Resour Crop Evol 53(5):1145–1152. https://doi.org/10.1007/s10722-005-1304-y
Lusas EW, Jividen GM (1987) Glandless cottonseed: a review of the west 25 years of processing and utilization research. J Am Oil Chem Soc 64:839–854. https://doi.org/10.3389/fpls.2021.753426
Mishra KK, Fougat RS (2014) Genetic diversity and molecular analysis among cotton genotypes by EST-SSR markers. Int J Environ 7:439–450.
Palle SR, Campbell LM, Pandeya D, Puckhaber L, Tollack LK (2013) RNAi-mediated ultra-low gossypol cottonseed trait: performance of transgenic lines under field conditions. Plant Biotechnol J 11:296–304
Rahman M, Shaheen T, Tabbasam N, Iqbal MA, Ashraf M (2012) Cotton genetic resources. A review. Agron Sustain Dev 32:419–432. https://doi.org/10.1007/s13593-011-0051-z
Roldan-Ruiz I, Dendauw J, Vanbockstaele E, Depicker A, De Loose M (2000) AFLP markers reveal high polymorphism rates in ryegrass (Lolium spp.). Mol Bred 6:125–134
Sapkal DR, Sutar SR, Thakre PB, Patil BR, Paterson AH, Waghmare VN (2011) Genetic diversity analysis of maintainer and restorer accessions in upland cotton (Gossypium hirsutum L.). J Plant Biochem Biotechnol 20:20–28. https://doi.org/10.1007/s13562-010-0020-7
Sarode DK, Sharma KM, Shinde NA, Gaikwad AR, Chavhan RL, Pimpale PA (2021) Molecular characterization of cotton (Gossypium hirsutum L.) genotypes using SSR markers for fiber quality traits. Int J Curr Microbiol and Appl Sci 10(03):1748–1759. https://doi.org/10.20546/ijcmas.2021.1003.218
Singh BD. Plant breeding principles and methods, Chapter 30. 2021. pp. 539–540.
Su JJ, Li LB, Pang CY (2016) Two genomic regions associated with fiber quality traits in Chinese upland cotton under apparent breeding selection. Sci Rep 7(6):38496. https://doi.org/10.1038/srep38496
Sun Z, Wang X, Liu Z, Gu Q, Zhang Y, Li Z, Ke H, Yang J, Wu J, Wu L, Zhang G, Zhang C, Ma Z (2017) Genome wide association study discovered genetic variation and candidate genes of Fiber quality traits in Gossypium hirsutum L. Plant Biotechnol J 15:982–996
Sundar R, Patil SS, Ranganath HM, Srivalli PR, Manjula SM, Pranesh KJ (2014) Genetic diversity analysis in cotton (Gossypium hirsutum L.) based on morphological traits and microsatellite markers. Int J Basic Appl Biol 1(1):19–22
Tang S, Teng Z, Zhai T, Fang X, Liu F, Liu D, Zhang J, Liu D, Wang S, Zhang K, Shao Q, Tan Z, Paterson AH, Zhang Z (2015) Construction of genetic map and QTL analysis of fiber quality traits for upland cotton (Gossypium hirsutum L.). Euphytica 201:195–213. https://doi.org/10.3389/fpls.2018.00225
Author Information
College of Agriculture, Pune, India