Molecular identification and evaluation of yield and grain quality of buckwheat varieties in Vietnam
*Article not assigned to an issue yet
Ta Linh Hong, Pham Duc Thanh, Le Yen Thi Hoang, Tran Trung Duc, Pham Ha Thi Thu, Nguyen Trung Thanh, Tran Tram Bao, Vu Tao Xuan, Le Yen Thi Hoang, Tran Trung Duc
Research Articles | Published: 04 December, 2025
First Page: 0
Last Page: 0
Views: 82
Keywords: Buckwheat, n Fagopyrum esculentumn , Molecular identification, Quality, Yield
Abstract
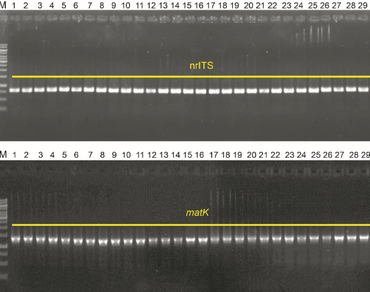
Buckwheat is widely recognized as a pseudocereal with considerable nutritional and agronomic importance. In Vietnam’s northern provinces, this crop is cultivated extensively for human consumption, livestock feed, and landscape enhancement, with a focus on tourism development. The current study aimed to molecularly identify buckwheat accessions from the northern mountainous regions of Vietnam and assess the seed quality of selected high-yielding varieties. A total of 32 buckwheat accessions were collected from local communes with diverse cultures and indigenous knowledge in agricultural practice across the region. Molecular authentication was conducted using the nuclear ribosomal internal transcribed spacer (nrITS) and the plastid matK sequences. Maximum likelihood phylogenetic inference revealed the robust affinities of the collected samples with reference F. esculentum, which were supported by high bootstrap values of 0.999 and 1.000 for the nrITS and matK sequences, respectively. Among these, five accessions—designated TGM28, TGM29, TGM30, TGM31, and TGM32—showed particularly high grain yields, with recorded outputs of 11.67, 9.33, 9.20, 8.40, and 8.67 quintals per hectare, respectively. Subsequent grain quality analysis of these five high-yielding accessions indicated that they are excellent nutritional sources, primarily composed of carbohydrates, proteins, fats, and fiber, and are notably gluten-free.
References
Álvarez I, Wendel JF (2003) Ribosomal ITS sequences and plant phylogenetic inference. Mol Phylogenet Evol 29(3):417–434. https://doi.org/10.1016/S1055-7903(03)00208-2
Aubert L, Decamps C, Jacquemin G, Quinet M (2021a) Comparison of plant morphology, yield and nutritional quality of Fagopyrum esculentum and Fagopyrum tataricum grown under field conditions in Belgium. Plants 10(2):258. https://doi.org/10.3390/plants10020258
Aubert L, Konrádová D, Barris S, Quinet M (2021b) Different drought resistance mechanisms between two buckwheat species Fagopyrum esculentum and Fagopyrum tataricum. Physiol Plant 172(2):577–586. https://doi.org/10.1111/ppl.13248
Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ (1995) The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann Missouri Bot Gard 82(2):247–277. https://doi.org/10.2307/2399880
Bhat S, Nazir M, Zargar SA, Naik S, Dar WA, Bhat BA, Mahajan R, Ganai BA, Sofi PA, Zargar SM (2022) In-depth morphological assessment revealed significant genetic variability in common buckwheat (Fagopyrum esculentum) and tartary buckwheat (Fagopyrum tataricum) germplasm. Plant Genet Resour 20(6):417–424. https://doi.org/10.1017/S1479262123000321
Campbell CG (1997) Buckwheat: Fagopyrum esculentum Moench, vol 19. Bioversity International, Roma
Cheng C, Fan Y, Tang Y, Zhang K, Joshi DC, Jha R, Janovská D, Meglič V, Yan M, Zhou M (2020) Fagopyrum esculentum ssp. ancestrale-a hybrid species between diploid F. cymosum and F. esculentum. Front Plant Sci 11:1073. https://doi.org/10.3389/fpls.2020.01073
Cho KS, Hong SY, Yun BK, Won HS, Yoon YH, Kwon KB (2017) Application of InDel markers based on the chloroplast genome sequences for authentication and traceability of Tartary and common buckwheat. Czech J Food Sci 35(2):122–130. https://doi.org/10.17221/116/2016-CJFS
Christa K, Soral-Śmietana M (2008) Buckwheat grains and buckwheat products–nutritional and prophylactic value of their components—a review. Czech J Food Sci 26(3):153–162. https://doi.org/10.17221/1602-CJFS
Fan Y, Jin YN, Ding M, Tang Y, Cheng J, Zhang K, Zhou M (2021) The complete chloroplast genome sequences of eight Fagopyrum species: insights into genome evolution and phylogenetic relationships. Front Plant Sci 12:799904. https://doi.org/10.3389/fpls.2021.799904
Gunkova PI, Buchilina AS, Ishevskiy AL, Pchelina EA, Nikolaev IA (2021) Chemical composition of buckwheat groats from various Russian manufacturers. IOP Conf Ser Earth Environ Sci 852:1–4. https://doi.org/10.1088/1755-1315/852/1/012036
Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, Ratnasingham S, van-der-Bank M, Chase MW, Cowan RS, Erickson DL, Fazekas AJ, Graham SW, James KE, Kim KJ, Kress WJ, Schneider H, van-AlphenStahl J, Barrett SCH, van-den-Berg C, Bogarin D, Burgess KS, Cameron KM, Carine M, Chacón J, Clark A, Clarkson JJ, Conrad F, Devey DS, Ford CS, Hedderson TAJ, Hollingsworth ML, Husband BC, Kelly LJ, Kesanakurti PR, Kim JS, Kim YD, Lahaye R, Lee HL, Long DG, Madriñán S, Maurin O, Meusnier I, Newmaster SG, Park CW, Percy DM, Petersen G, Richardson JE, Salazar GA, Savolainen V, Seberg O, Wilkinson MJ, Yi DK and Little DP (2009) A DNA barcode for land plants. Proc Natl Acad Sci USA 106(31):12794–12797. https://doi.org/10.1073/pnas.0905845106
Huang XY, Zeller FJ, Huang KF, Shi TX, Chen QF (2014) Variation of major minerals and trace elements in seeds of tartary buckwheat (Fagopyrum tataricum Gaertn.). Genet Resour Crop Evol 61:567–577. https://doi.org/10.1007/s10722-013-0057-2
Kalyaanamoorthy S, Minh BQ, Wong TK, Von-Haeseler A, Jermiin LS (2017) ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14(6):587–589. https://doi.org/10.1038/nmeth.4285
Knicky M, Fogelberg F, Galia Z (2024) Yield and protein composition in seeds of four buckwheat (Fagopyrum sp.) cultivars cropped in Sweden. Agric Food Sci 33:261–267. https://doi.org/10.23986/afsci.147755
Kolarić L, Popović V, Živanović L, Ljubičić N, Stevanović P, Šarčević Todosijević L, Simić D, Ikanović J (2021) Buckwheat yield traits response as influenced by row spacing, nitrogen, phosphorus, and potassium management. Agronomy 11(12):2371. https://doi.org/10.3390/agronomy11122371
Li C, Xie Z, Wang Y, Lu W, Yin G, Sun D, Ren C, Wang L (2019) Correlation and genetic analysis of seed shell thickness and yield factors in Tartary buckwheat (Fagopyrum tataricum (L.) Gaertn.). Breed Sci 69(3):464–470. https://doi.org/10.1270/jsbbs.18081
Luitel RD, Siwakoti M, Jha KP, Jha KA, Krakauer N (2017) An overview: distribution, production, and diversity of local landraces of buckwheat in Nepal. Advances in Agriculture 2017(1):1–6. https://doi.org/10.1155/2017/2738045
Luthar Z, Zhou M, Golob A, Germ M (2020) Breeding buckwheat for increased levels and improved quality of protein. Plants 10(1):1–12. https://doi.org/10.3390/plants10010014
Madeira F, Pearce M, Tivey AR, Basutkar P, Lee J, Edbali O, Madhusoodanan N, Kolesnikov A, Lopez R (2022) Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res 50(W1):276–279. https://doi.org/10.1093/nar/gkac240
Min D, Shi W, Dehshiri MM, Gou Y, Li W, Zhang K, Zhou M, Li B (2023) The molecular phylogenetic position of Harpagocarpus (Polygonaceae) sheds new light on the infrageneric classification of Fagopyrum. PhytoKeys 220:109–126. https://doi.org/10.3897/phytokeys.220.97667
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von-Haeseler A, Lanfear R (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37(5):1530–1534. https://doi.org/10.1093/molbev/msaa015
Neacsu M, De-Lima Sampaio S, Hayes HE, Duncan GJ, Vaughan NJ, Russell WR, Raikos V (2022) Nutritional content, phytochemical profiling, and physical properties of buckwheat (Fagopyrum esculentum) seeds for promotion of dietary and food ingredient biodiversity. Crops 2(3):287–305. https://doi.org/10.3390/crops2030021
Ohsako T, Li C (2020) Classification and systematics of the Fagopyrum species. Breed Sci 70(1):93–100. https://doi.org/10.1270/jsbbs.19028
Rana J (2004) Buckwheat genetic resources management in India. In: Proc 9th Int Symp Buckwheat, Prague, 271–282
Selvaraj D, Park JI, Chung MY, Cho YG, Ramalingam S, Nou IS (2013) Utility of DNA barcoding for plant biodiversity conservation. Plant Breed Biotechnol 1(4):320–332. https://doi.org/10.9787/PBB.2013.1.4.320
Smillie T, Khan I (2010) A comprehensive approach to identifying and authenticating botanical products. Clin Pharmacol Ther 87(2):175–186. https://doi.org/10.1038/clpt.2009.287
Sofi SA, Ahmed N, Farooq A, Rafiq S, Zargar SM, Kamran F, Dar TA, Mir SA, Dar BN, Khaneghah AM (2023) Nutritional and bioactive characteristics of buckwheat, and its potential for developing gluten-free products: an updated overview. Food Sci Nutr 11(5):2256–2276. https://doi.org/10.1002/fsn3.3166
Sohn HB, Kim SJ, Hong SY, Park SG, Oh DH, Lee S, Nam HY, Nam JH, Kim YH (2021) Development of 50 InDel-based barcode system for genetic identification of tartary buckwheat resources. PLoS ONE 16(6):1–20. https://doi.org/10.1371/journal.pone.0250786
Sonawane S, Shams R, Dash KK, Patil V, Pandey VK, Dar AH (2024) Nutritional profile, bioactive properties and potential health benefits of buckwheat: a review. eFood 5(4):1–17. https://doi.org/10.1002/efd2.171
Wen J, Zimmer EA (1996) Phylogeny and biogeography of Panax L. (the ginseng genus, Araliaceae): inferences from ITS sequences of nuclear ribosomal DNA. Mol Phylogenet Evol 6(2):167–177. https://doi.org/10.1006/mpev.1996.0069
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13(1):134. https://doi.org/10.1186/1471-2105-13-134
Zhou Q, He P, Tang J, Huang K, Huang X (2023) Increasing planting density can improve the yield of Tartary buckwheat. Front Plant Sci 14:1–10. https://doi.org/10.3389/fpls.2023.1313181
Author Information
Vietnam Academy of Agricultural Sciences, Ministry of Agriculture and Environment, Hanoi, Vietnam