Morphometry of Mimosa caesalpiniifolia Benth. (Fabaceae) seeds through digital image analysis in a genetic divergence study
*Article not assigned to an issue yet
da Silva Santos Paulo César, de Sousa Moema Barbosa, da Silva Carlos Luiz, Nonato Erika Rayra Lima, de Freitas Eliane Cristina Sampaio, Santos Marcone Moreira, Gallo Ricardo
Research Articles | Published: 16 October, 2025
First Page: 0
Last Page: 0
Views: 2
Keywords: Sabiá, Morphogenetic studies, Genotypic variation, Digital technology
Abstract
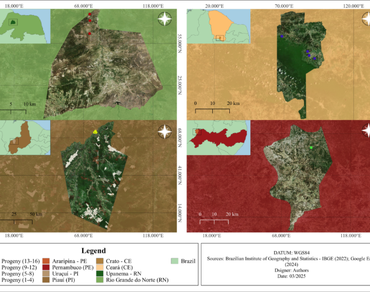
Mimosa caesalpiniifolia Benth. (Fabaceae), or sabiá, is native to the Caatinga and essential for degraded ecosystems, as it fixes nitrogen and favors regeneration. Its seeds have morphometric characteristics that are important for reproductive ecology and conservation. However, measuring them accurately is challenging due to natural variability and the need for advanced methods such as digital images and statistical analysis. Therefore, the objective of this study was to evaluate the potential of digital image analysis in seeds and morphometrically characterize M. caesalpiniifolia seeds with the aim of applying genetic divergence in traits for mother tree selection. For this purpose, four progenies from four different origins in the states of Ceará, Paraíba, Pernambuco, and Rio Grande do Norte were selected, totaling 16 progenies. The genetic diversity of the seeds was evaluated using two methods: digital caliper and ImageJ® program. Subsequently, Pearson’s correlation analyses between the methods, as well as phenotypic and genotypic correlation analyses between morphometric variables, multivariate, and genetic analyses were conducted. Genotypic variance was significant (p < 0.05) for most traits, with high heritability values (H2 > 0.60) for seed area, perimeter, width, and length, indicating strong genetic control. The digital image processing proved to be efficient in discriminating the morphometric differences among the studied origins, which exhibited variations in morphometric aspects due to the genetic differences of the species, aiding in the differentiation of genotypes. This study demonstrates the potential of digital image analysis as a rapid and precise tool for the morphometric characterization of M. caesalpiniifolia seeds, enabling the identification of genetic divergence and facilitating the selection of seed-producing genotypes for conservation and breeding efforts.
References
Alves EU, Bruno RLA, Alves AU, Cardoso EA, Galindo EA, Braga Junior JM (2007) Germinação e biometria de frutos e sementes de Bauhinia divaricata L. Sitientibus Série Ciências Biológicas 7:193–198
Andrade DB, dos Oliveira AS, Pinto CAG, de Pires RMO, Oliveira AS, Silva MA, Carvalho MLM (2016) Detection of green seeds in soybean lots by the seed analysis system (SAS). Int J Currss Res 8:26462–26465
Andrew SM, Kombo SA, Chamshama SAO (2021) Diversity in fruit and seed morphology of wooden banana (Entandrophragma bussei Harms ex Engl.) populations in Tanzania. Trees Forests People 5:100095. https://doi.org/10.1016/J.TFP.2021.100095
Araújo FDS, Felix FC, Ferrari CDS, Vieira FDA, Pacheco MV (2020) Seed quality and genetic diversity of a cultivated population of Mimosa caesalpiniifolia Benth. Rev Caatinga 33:1000–1006. https://doi.org/10.1590/1983-21252020v33n415rc
Baek J, Lee E, Kim N, Kim SL, Choi I, Ji H, Chung YS, Choi MS, Moon JK, Kim KH (2020) High throughput phenotyping for various traits on soybean seeds using image analysis. Sensors (Switzerland) 20:1–9. https://doi.org/10.3390/s20010248
Balestre M, von Pinho RG, de Souza Junior CL, de Bueno Filho JSS (2012) Bayesian mapping of multiple traits in maize: the importance of pleiotropic effects in studying the inheritance of quantitative traits. Theor Appl Genet. https://doi.org/10.1007/s00122-012-1847-1
Benavides R, Carvalho B, Bastias CC, López-Quiroga D, Mas A, Cavers S, Valladares F (2021) The gentree leaf collection: inter-and intraspecific leaf variation in seven forest tree species in Europe. Glob Ecol Biogeogr 30(3):590–597. https://doi.org/10.1111/geb.13239
Borborema LDA, Junior EDOF, de Sousa Gonçalves RJ, dos Santos JS, dos Santos Rodrigues APM, de MendonçaJúnior AF (2020) Diversidade genética entre acessos de feijão-guandu por meio de caracteres morfoagronômicos em região do cariri paraibano. Revista Verde De Agroecologia e Desenvolvimento Sustentável 15(2):133–138. https://doi.org/10.18378/rvads.v15i2.8252
Callegari-Jacques, S.M., 2003. Bioestatistica: princípios e aplicações. Artmed, Porto Alegre.
Costa MF, Lopes ACdeA, Gomes RLF, Araújo ASF, Zucchi MI, Pinheiro JB, Valente SEdosS (2016) Caracterização e divergência genética de populações de Casearia grandiflora no cerrado Piauiense. Florest Amb 23:387–396. https://doi.org/10.1590/2179-8087.007115
Cruz CD (2013) GENES—a software package for analysis in experimental statistics and quantitative genetics. Acta Sci Agron 35:271–276
Cruz CD, Regazzi AJ, Carneiro PCS (2014) Modelos biométricos aplicados ao melhoramento genético, 3rd edn. Editora UFV, Viçosa
Cruz, C.D., Regazzi, A.J., Carneiro, P.C.S., 2012. Modelos biométricos aplicados ao melhoramento genético. Editora UFV, Viçosa.
da Silva APM, Leitão LRG, Araújo LBR, Cunha Neto J, Bertini CHCDM (2021) Genetic progress and early selection of juvenile physic nut genotypes. Nativa. https://doi.org/10.31413/nativa.v9i5.11542
Dayrell RL, Ott T, Horrocks T, Poschlod P (2023) Automated extraction of seed morphological traits from images. Methods Ecol Evol 14(7):1708–1718. https://doi.org/10.1111/2041-210X.14127
Felix FC, Medeiros JAD, Ferrari CS, Vieira FA, Pacheco MV (2020) Biometry of Pityrocarpa moniliformis seeds using digital imaging: implications for studies of genetic divergence. Rev Bras Cienc Agrar 15:1–8. https://doi.org/10.5039/agraria.v15i1a6128
Gao T, Chandran AKN, Paul P, Walia H, Yu H (2021) Hyperseed: an end-to-end method to process hyperspectral images of seeds. Sensors 21(24):8184. https://doi.org/10.3390/s21248184
Gois IB, Ferreira RA, Silva-Mann R (2018) Variabilidade Genética em Populações Naturais de Cassia grandis L. f. Florest Amb 25:1–10. https://doi.org/10.1590/2179-8087.160309
Gonçalves GV, Andrade FR, Hur B, Junior M, Schossler R, Lenza E, Marimon BS (2013) Biometria de frutos e sementes de mangaba (Hancornia speciosa Gomes) em vegetação natural na região leste de Mato Grosso, Brasil. Rev Cienc Agrar 36:31–40
Granitto PM, Verdes PF, Ceccatto HA (2005) Large-scale investigation of weed seed identification by machine vision. Comput Electron Agric 47:15–24. https://doi.org/10.1016/j.compag.2004.10.003
https://ggplot2.tidyverse.org.
Jiménez-Lópes F, Talavera M, Ortiz MA (2024) Morphological and genetic characterization of genotypic diversity using morphological descriptors in Mimosa caesalpiniifolia Benth. germplasm. Ind Crops Prod. https://doi.org/10.1002/tax.13248
Kapadia VN, Sasidharan N, Kalyanrao P (2017) Seed image analysis and its application in seed science research. Adv Biotechnol Microbiol. https://doi.org/10.19080/aibm.2017.07.555709
Kassambara, A. and Mundt, F. (2020) Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R Package Version 1.0.7. https://CRAN.R-project.org/package=factoextra
Krause W, Viana A, Cavalcante N, Ambrósio M, Santos E, Vieira H (2017) Digital phenotyping for quantification of genetic diversity in inbred guava (Psidium guajava) families. Genet Mol Res 16:1–11. https://doi.org/10.4238/gmr16019474
Leão NVM, Felipe SHS, Emídio-Silva C, Moraes ACS, Shimizu ESC, Gallo R, Freitas ADD, Kato OK (2018) Morphometric diversity between fruits and seeds of mahogany trees (Swietenia macrophylla King.) from Parakanã Indigenous Land Pará State Brazil. Aust J Crop Sci. https://doi.org/10.21475/ajcs.18.12.03.pne879
Lestari DA, Faya AKN (2024) Seed Conservation of Anaxagorea luzonensis A. Gray (Annonaceae) through storage behaviour and morphology. J Trop Biodivers Biotechnol. https://doi.org/10.22146/jtbb.83147
Loveless MD, Hamrick JL (1984) Ecological determinants of genetic structure in plant populations. Annu Rev Ecol Syst 15:65–95. https://doi.org/10.1146/ANNUREV.ES.15.110184.000433
Margapuri V, Courtney C, Neilsen M (2021) Image processing for high-throughput phenotyping of seeds. EPiC Series in Computing 75:69–79
Matsuda O, Tanaka A, Fujita T, Iba K (2012) Hyperspectral imaging techniques for rapid identification of Arabidopsis mutants with altered leaf pigment status. Plant Cell Physiol 53:1154–1170. https://doi.org/10.1093/pcp/pcs043
Medeiros AD, Pereira MD, Silva JA (2018) Processamento digital de imagens na determinação do vigor de sementes de milho. Rev Bras Cienc Agrar 13:1–7. https://doi.org/10.5039/agraria.v13i3a5540
Moraes MC, Mengarda LHG, Canal GB, Pereira PM, Ferreira A, Ferreira MFS (2020) Diversidade genética de matrizes e progênies de Euterpe edulis Mart. em área manejada e em populações naturais por marcadores microssatélites. Ciênc Florest 30:583–594. https://doi.org/10.5902/1980509837647
Musaev FB, Beletskiy SL, Potrakhov NN (2021) Development of computer program for automatic X-ray analysis of quality of vegetable seeds. Plant Biol Hortic Theory Innov 160:86–95
Pinheiro RD, Ferreira EJL, Marques RA, Carvalho HP, Barros Q, Silva FAF (2024) Analysis of genotypic diversity using morphological descriptors in seeds of Tabebuia rosea (Bignoniaceae). Ciência Agrotecn. https://doi.org/10.1590/1413-7054202448007324
Pinto MS, Damasceno Junior PC, Oliveira LC, Machado AFdeF, Souza MAA, Muniz DR, Dias LAdosS (2018) Diversity between Jatropha curcas L. accessions based on oil traits and X-ray digital images analysis from it seeds. Crop Breed Appl Biotechnol 18:292–300. https://doi.org/10.1590/1984-70332018v18n3a43
R Core Team (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/.
Rabieyan E, Bihamta M, EsmaeilzadehMoghaddam M, Mohammadi V, Alipour H (2022) Morpho-colorimetric seed traits for the discrimination, classification and prediction of yield in wheat genotypes under rainfed and well-watered conditions. Crop Pasture Sci 74(3):294–311. https://doi.org/10.1071/CP22127
Resende MDv (2016) Software selegen-REML/BLUP: a useful tool for plant breeding. Crop Breed Appl Biotechnol 16:330–339
Resende MDV, Duarte JB (2007) Precisão e controle de qualidade em experimentos de avaliação de cultivares. Pesqui Agropecu Trop 37:182–194
Resende, M.D.V., 2007. Matemática e estatística na análise de experimentos e no melhoramento genético. Embrapa Florestas, Colombo.
Rocha JDL, Guilherme F, Rocha DI, Pereira KAR, Coelho CP, Souza LF (2022) Morphometry of fruits and pyrenes in two morphotypes and populations of Butia purpurascens Glassman (Arecaceae). Cienc Rural 52(8):e20210303. https://doi.org/10.1590/0103-8478cr20210303
Roscher R, Herzog K, Kunkel A, Kicherer A, Töpfer R, Förstner W (2014) Automated image analysis framework for high-throughput determination of grapevine berry sizes using conditional random fields. Comput Electron Agric 100:148–158. https://doi.org/10.1016/j.compag.2013.11.008
Rossi FS, Aparecida A, Rossi B, Dardengo J, Silva ML (2014) Diversidade genética em populações naturais de Mauritia flexuosa L. f. (Arecaceae) com uso de marcadores ISSR. Scientia for 42:631–639
Rossi AAB, da Silva MDSA, Arenas-de-Souza MD, Zortéa KÉM, Della Giustina L, da Rocha VD (2017) Diversidade genética de duas espécies de Theobroma L. em um fragmento florestal no Portal da Amazônia MT Brasil. Nativa. https://doi.org/10.5935/2318-7670.v05nespa08
Rotili EA, Cancellier LL, Dotto MA, Peluzio JM, de Carvalho EV (2012) Divergência genética em genótipos de milho, no Estado do Tocantins. Rev Cienc Agron 43:516–521
RoveriNeto A, Paula RC (2017) Variabilidade entre árvores matrizes de Ceiba speciosa St. Hil para características de frutos e sementes. Revista Ciencia Agron 48:318–327. https://doi.org/10.5935/1806-6690.20170037
Sau S, Ucchesu M, D’hallewin, G., Bacchetta, G., (2019) Potential use of seed morpho-colourimetric analysis for Sardinian apple cultivar characterisation. Comput Electron Agric. https://doi.org/10.1016/j.compag.2019.04.027
Silva VN, Sarmento MB, Silveira AC, Silva CS, Cicero SM (2013) Avaliação da morfologia interna de sementes de Acca sellowiana O. Berg por meio de análise de imagens. Rev Bras Frutic 35:1158–1169. https://doi.org/10.1590/S0100-29452013000400027
Silva RMda, Cardoso AD, Dutra FV, Morais OM (2017) Aspectos biométricos de frutos e sementes de Caesalpinia ferrea Mart. ex Tul. provenientes do semiárido baiano. Rev Agric Neotrop 4:85–91. https://doi.org/10.32404/rean.v4i3.1427
Singh D (1981) The relative importance of characters affecting genetic divergence. Indian J Genet Plant Breed 41:237–245
Soares JM, Medeiros ADD, Pinheiro DT, Rosas JTF, Silva LJD, Machado DLM, Dias DCFDS (2023) Low-cost system for multispectral image acquisition and its applicability to analysis of the physiological potential of soybean seeds. Acta Sci Agron 45:e57060. https://doi.org/10.4025/actasciagron.v45i1.57060
Srivastava A, Gupta S, Shanker K, Gupta N, Gupta AK, Lal RK (2020) Genetic diversity in Indian poppy (P. somniferum L.) germplasm using multivariate and SCoT marker analyses. Ind Crops Prod 144:112050. https://doi.org/10.1016/J.INDCROP.2019.112050
Ucchesu M, Orrù M, Grillo O, Venora G, Paglietti G, Ardu A, Bacchetta G (2016) Predictive method for correct identification of archaeological charred grape seeds: support for advances in knowledge of grape domestication process. PLoS ONE. https://doi.org/10.1371/journal.pone.0149814
Vale AMPG, Ucchesu M, Di Ruberto C, Loddo A, Soares JM, Bacchetta G (2020) A new automatic approach to seed image analysis: from acquisition to segmentation. arXiv Prep. https://doi.org/10.48550/arXiv.2012.06414
Vencovsky, R., Barriga, P., 1992. Genética Biométrica no Melhoramento. SBG, Ribeirão Preto.
Watson A, Hickey LT, Christopher J, Rutkoski J, Poland J, Hayes BJ (2019) Multivariate genomic selection and potential of rapid indirect selection with speed breeding in Spring wheat. Crop Sci 59:1945–1959. https://doi.org/10.2135/CROPSCI2018.12.0757
Wei, T., Simko, V. (2017). R package “corrplot”: Visualization of a Correlation Matrix. https://github.com/taiyun/corrplot
Wickham H (2016) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York
Wiwart M, Kurasiak-Popowska D, Suchowilska E, Wachowska U, Stuper-Szablewska K (2019) Variation in the morphometric parameters of seeds of spring and winter genotypes of Camelina sativa (L.) Crantz. Ind Crops Prod 139:1–7. https://doi.org/10.1016/j.indcrop.2019.111571
Wright S (1978) Evolution and genetics of populations. University of Chicago, Chicago
Xavier MVB, Fonseca APM, de Almeida ES, de Oliveira PVA (2020) Manejo do Oncideres ocularis Thomson em Mimosa caesalpiniifolia Benth. Res Soc Dev 9(7):e787974601. https://doi.org/10.33448/rsd-v9i7.4601
Ferreira CBB, Lopes MTG, Lopes R, Cunha RNV, Moreira DA, Barros WS, Matiello RR (2012) Diversidade genética molecular de progênies de dendezeiro tipo Dura de origem Deli. Pesquisa Agropecuária Brasileira 47(3):378–384. https://doi.org/10.1590/S0100-204X2012000300009
IUCN (2016) INTERNATIONAL UNION FOR CONSERVATION OF NATURE. World conservation monitoring centre. IUCN Red List of Threatened Species. Versãão 2011.2, IUCN. Disponíível em: . Acesso em: 18 Jan. 2016. Version 2016.2, IUCN. Accessed: Jan. 18, 2019
Author Information
Fedral Rural University of Pernambuco, Recife, Brazil