Principal component and cluster analysis-assisted selection of potato (Solanum tuberosum. L) genotypes
*Article not assigned to an issue yet
K R Aishwarya, Singh Dhirendra, Pradeep S D, Mehta Tanvi, Singh Khyati, Sudesh, Nitwal Renu, Singh J P.
Research Articles | Published: 24 July, 2025
First Page: 0
Last Page: 0
Views: 111
Keywords: Cluster analysis, Principal component analysis, Potato, Yield, Yield related traits
Abstract
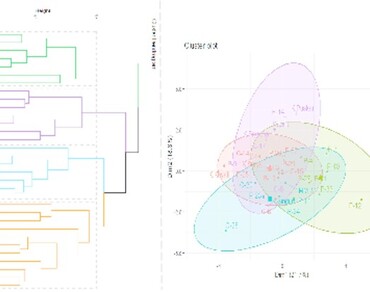
Potato (Solanum tuberosum L.) is a globally vital food crop and a rich source of nutrients for human populations, playing a key role in food security. This study aimed to identify superior elite potato genotypes using hierarchical cluster analysis (HCA) and principal component analysis (PCA) for fourteen qualitative and quantitative traits. Hierarchical clustering grouped genotypes into four distinct clusters, with Cluster I containing the maximum number (11) of genotypes showing high performance for tuber growth and quality traits. The intra-cluster and inter-cluster distances ranged from 5.73 (Cluster I) to 3.92 (Cluster III) and 6.81 (between Clusters II and IV) to 4.98 (Clusters I and III), respectively, indicating the presence of significant genetic variation among the studied genotypes. The PCA analysis revealed six principal components (PCs) with eigenvalues exceeding one explained 78.43% of the total variance, with plant height at 60 days after planting, tuber girth, tuber length, number of tubers per plant, the average weight of tuber per plant, specific gravity of tuber and tuber yield per plot contributed more to PC1 and PC2 and indicating their positive influence on tuber yield. Based on statistical analysis, the genotypes in Cluster III and Cluster IV can be considered the most suitable parents for improving yield related traits. Considering HCA and PCA of qualitative and quantitative traits, the genotypes C-6, C-14, C-17, C-20, C-28, P-9, P-23, and P-25 may be selected as promising lines for selection as optimal parents in future breeding programs focused on improving yield and nutritional quality.
References
Alam Z, Khan MAH, Hossain MI, Karim MR, Saif HB, Mustakim AM, Akter S (2024) Genetic variability and diversity analysis for some agronomic traits of a sweet potato (Ipomoea Batatas L.) collection: insights for breeding superior genotypes. Heliyon 10:e38616. https://doi.org/10.1016/j.heliyon.2024.e38616
Asghari M, Kalantar M, Hassanpanah D, Dehghani Zahedani M (2022) Genetic varieties of the obtained hybrids from the potato (Solanum tuberosum L.) tuber intersections in the spring using factor and cluster analysis. J Veg Sci 10:163–179. https://doi.org/10.22034/iuvs.2021.522522.1142
Azam MG, Hossain MA, Sarker U, Alam AM, Nair RM, Roychowdhury R, Golokhvast KS (2023) Genetic analyses of mungbean [Vigna radiata (L.) Wilczek] breeding traits for selecting superior genotype(s) using multivariate and multi-traits indexing approaches. Plants 12:1984. https://doi.org/10.3390/plants12101984
Beheshtizadeh H, Resaaie AH, Resaaie AM, Ghandi A (2013) Genetic variability assessment in bread wheat (Triticum aestivum L.) cultivars using multivariate statistical analysis. Int J Farming Allied Sci 2:520–523
Bodeddula JR, Mandal R, Nag S, Nath S, Chakraborty M, Hijam L, Roy SK (2021) Evaluation and characterization of local potato cultivars collected from the Northern part of West Bengal based on phenology. Int J Ecol Environ Sci 47:287–301
FAOSTAT (2019) Food and Agriculture Organization of the United Nations-Statistic Division. https://www.fao.org/faostat/en/#data (Accessed 23 Nov 2021)
FAOSTAT (2022) FAOSTAT database. https://www.fao.org/faostat/en/#data/QC (Accessed 19 Feb 2022)
Gadissa F, Tesfaye K, Dagne K, Geleta M (2020) Morphological traits based genetic diversity assessment of Ethiopian potato [Plectranthus edulis (Vatke) Agnew] populations. Genet Resour Crop Evol 67:809–829. https://doi.org/10.1007/s10722-020-00884-1
Gowda KH, Amarananjundeswara H, Fakrudin B, Vasudeva KR, Doddabasappa B, Kattegoudar J (2025) Biochemical characterization, sensory evaluation, instrumental colour profiling and selection of nutritional-rich genotypes through hierarchical cluster analysis, multivariate analysis and multi-trait genotype ideotype – distance index in potato (Solanum tuberosum L). Potato Res 1–37. https://doi.org/10.1007/s11540-025-0951-3
Gutiérrez-Valencia J, Slotte T (2022) Potato genomes pave the way to crop improvement. Curr Opin Plant Biol 66:102174. https://doi.org/10.1016/j.pbi.2022.102174
Hossain MM, Kaium MA, Al Amin M, Ali TB, Jahan N, Uddin MN (2023) Evaluation of genetic divergence in various potato genotypes grown in Bangladesh through different traits assessment. Am J Plant Sci 14:1235–1247. https://doi.org/10.4236/ajps.2023.1411087
Kashyap R, Rangare NR, Rangare SB (2021) Evaluation of potato (Solanum tuberosum L.) germplasm for some important morphological traits using multivariate analysis. Pharma Innov J 10:180–185
Kassambara A, Mundt F (2019) Factoextra: Extract and visualize the results of multivariate data analyses. R Foundation for Statistical Computing, Vienna, Austria. https://doi.org/10.32614/cran.package.factoextra
Kaur S, Aggarwal P, Babbar N (2023) Evaluating progress of Indian potato processing industry: an updated review. Potato Res 66:945–964. https://doi.org/10.1007/s11540-023-0960-5
Khan MA, Yousaf MW, Ahmed HGM, Fatima N, Alam B (2024) Assessing genetic diversity for some Pakistani bread wheat (Triticum aestivum L.) genotypes under drought stress based on yield traits. Genet Resour Crop Evol 1–11. https://doi.org/10.1007/s10722-024-01523-7
Kolde R (2018) pheatmap: Pretty Heatmaps. R package version 1.0.12. https://github.com/raivokolde/pheatmap
Lê S, Josse J, Husson F (2008) FactoMineR: an R package for multivariate analysis. J Stat Softw 25:1–18. https://doi.org/10.18637/jss.v025.i01
Lee KJ, Sebastin R, Cho GT, Yoon M, Lee GA, Hyun DY (2021) Genetic diversity and population structure of potato germplasm in RDA-genebank: utilization for breeding and conservation. Plants 10:752. https://doi.org/10.3390/plants10040752
Luthra SK, Kumar V (2024) Potato genetic resources and their utilization in India. Indian J Plant Genet Resour 37:1–19. https://doi.org/10.61949/0976-1926.2024.v37i01.01
Maechler M, Rousseeuw PJ, Struyf A, Hubert M, Hornik K, Studer M, Roudier P, Gonzalez J, Kozlowski K, Schubert E et al (2019) Cluster: Cluster analysis basics and extensions. R Foundation for Statistical Computing, Vienna, Austria. https://cran.r-project.org/web/packages/cluster/cluster.pdf
Mallick SR, Quamruzzaman AKM, Hossain MA, Rahman MM, Hoque MA, Islam MR (2021) Diversity of potato varieties in Bangladesh. Eur J Agric Food Sci 3:56–61. https://doi.org/10.24018/ejfood.2021.3.3.281
Mishra T, Raigond P, Thakur N, Dutt S, Singh B (2020) Recent updates on healthy phytoconstituents in potato: A nutritional depository. Potato Res 63:323–343. https://doi.org/10.1007/s11540-020-0945-2
Murtagh F, Legendre P (2011) Ward’s hierarchical clustering method: Clustering criterion and agglomerative algorithm. arXiv:1111.6285
Nasiruddin M, Ali F, Islam AKM (2017) Genetic diversity in potato (Solanum tuberosum L.) genotypes grown in Bangladesh. Int Res J Biol Sci 6:1–8
Pandey J, Scheuring DC, Koym JW, Vales MI (2022) Genomic regions associated with tuber traits in tetraploid potatoes and identification of superior clones for breeding purposes. Front Plant Sci 13:952263. https://doi.org/10.3389/fpls.2022.952263
Pradhan AM, Sarkar KK, Konar A (2015) Analysis of genetic diversity in Indian potato genotypes by using principal component analysis. Electron J Plant Breed 6:782–786. https://doi.org/10.5958/0975-928X.2015.00098.9
Rangare SB, Rangare NR (2017) Classificatory analysis of potato (Solanum tuberosum L.) genotypes for yield and yield attributing traits. Pharma Innov J 6:94–96
Sanchez D, Allier A, Ben Sadoun S, Mary-Huard T, Bauland C, Palaffre C, Charcosset A (2024) Assessing the potential of genetic resource introduction into elite germplasm: A collaborative multiparental population for Flint maize. Theor Appl Genet 137:19. https://doi.org/10.1007/s00122-023-04356-5
Seid E, Mohammed W, Abebe T (2021) Genetic diversity assessment through cluster and principal component analysis in potato (Solanum tuberosum L.) genotypes for processing traits. Int J Food Sci Agric 5:440–447. https://doi.org/10.26855/ijfsa.2021.09.014
Tang Y, Horikoshi M, Li W (2016) Ggfortify: unified interface to visualize statistical results of popular R packages. R J 8:478–489. https://doi.org/10.32614/RJ-2016-060
R Development Core Team (2010) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.r-project.org/
Wickham H (2016) ggplot2: elegant graphics for data analysis. Springer-, New York. https://doi.org/10.1007/978-3-319-24277-4
Wickham H, Chang W, Henry L, Pedersen TL, Takahashi K, Wilke C, Woo K, Yutani H, van den Dunnington D (2025) ggplot2: Create elegant data visualisations using the grammar of graphics. R package version 3.5.2. https://doi.org/10.32614/CRAN.package.ggplot2
Wickham H, François R, Henry L, Müller K (2020) dplyr: A grammar of data manipulations. R package version 0.8.5. https://cran.r-project.org/package=dplyr
Xie RX, Zhang XC, Wu LK, Guo ZQ, Zhang GH, Yu BQ (2021) Main quality traits analysis and evaluation of potato germplasms. Mol Plant Breed 12:1–8. https://doi.org/10.5376/mpb.2021.12.0028
Author Information
Department of Vegetable Science, College of Agriculture, CCS HAU, Hisar, India