Transcriptome-wide identification and characterization of Musa circular RNAs in corm and root tissues of resistant and susceptible cultivars in response to Fusarium oxysporum f.sp. cubense race1 and TR4
*Article not assigned to an issue yet
Arumugam Chandrasekar, Chelliah Anuradha, Punchakkara Prashina Mol, Suthanthiram Backiyarani, Raman Thangavelu, Ramasamy Selvarajan
Research Articles | Published: 27 May, 2025
First Page: 0
Last Page: 0
Views: 433
Keywords: ceRNA network, circRNAs, Expression pattern, Fusarium wilt, Gene regulation, miRNA sponges, Non-coding RNA
Abstract
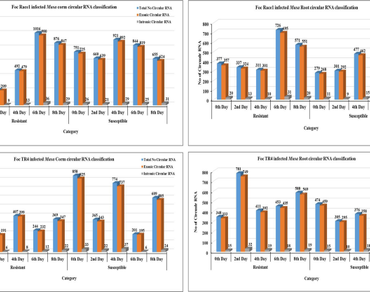
Fusarium wilt, caused by Fusarium oxysporum f.sp. cubense (Foc), poses a significant threat to banana crops. Circular RNAs (circRNAs), a newly discovered group of non-coding RNA, play a role in gene regulation. However, Musa spp. has not been explored for circRNA detection. This study presents the first identification of circRNAs in resistant and susceptible banana cultivars infected with Foc race 1 and TR4. A substantial number of circRNAs were identified from RNA-seq libraries of infected corm and root tissues. Differential expression analysis revealed specific circRNAs associated with Foc-resistant and susceptible cultivars. Functional annotation and pathway analysis highlighted involvement in secondary metabolite biosynthesis, Mitogen-activated protein kinases (MAPK) signalling pathway, and plant-pathogen interaction. Utilizing the differentially expressed circRNAs, we conducted a microRNA miRNA-mediated interaction analysis to identify potential sponge interactions. Subsequently, a comprehensive circRNA–miRNA–messenger RNA (mRNA) network was constructed, incorporating target genes directly or indirectly associated with plant resistance. This network sheds light on the intricate regulatory mechanisms underlying plant resistance to Fusarium wilt and provides valuable insights into the complex interplay between circRNAs, miRNAs, and mRNAs in banana defense responses.
References
Anuradha C, Chandrasekar A, Backiyarani S, Uma S (2022) MusaRgeneDB: an online comprehensive database for disease resistance genes in Musa spp. 3 Biotech 12:1–12. https://doi.org/10.1007/s13205-022-03285-1
Anuradha C, Chandrasekar A, Backiyarani S, Thangavelu R, Uma S, Selvarajan R (2024) Dataset from transcriptome profiling of Musa resistant and susceptible cultivars in response to Fusarium oxysporum f sp cubense race1 and TR4 challenges using Illumina NovaSeq. Data Br 52:109803. https://doi.org/10.1016/jdib2023109803
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Kadener S (2014) circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56(1):55–66. https://doi.org/10.1016/jmolcel201408019
Bai TT, Xie WB, Zhou PP, Wu ZL, Xiao WC, Zhou L, Li HP (2013) Transcriptome and expression profile analysis of highly resistant and susceptible banana roots challenged with Fusarium oxysporum f sp cubense tropical race 4. PLOS One 8(9):e73945. https://doi.org/10.1371/journalpone0073945
Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE (2006) Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18(4):1038–51. https://doi.org/10.1105/tpc105039982
Bazzini AA, Hopp HE, Beachy RN, Asurmendi S (2007) Infection and co-accumulation of tobacco mosaic virus proteins alter microRNA levels, correlating with symptom and plant development. Proc Natl Acad Sci USA 104(29):12157–12162. https://doi.org/10.1073/pnas0705114104
Bhavya C, Dayanandhi E, Sadashiva AT, Reddy MK, Ravishankar KV (2022) Identification of circular RNAs in resistant tomato genotype in response to ToLCBaV infection. Int J Hortic Sci 17(2):496–504. https://doi.org/10.24154/jhsv17i21520
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Bordoloi KS, Baruah PM, Agarwala N (2023) Identification of circular RNAs in tea plant during Helopeltis theivora infestation. Plant Stress 8:100150. https://doi.org/10.1016/jstress2023100150
Chandran V, Wang H, Gao F, Cao XL, Chen YP, Li GB, Wang WM (2019) miR396-OsGRFs module balances growth and rice blast disease-resistance. Front Plant Sci 9:1999. https://doi.org/10.3389/fpls201801999
Chelliah A, Arumugam C, Punchakkara PM, Suthanthiram B, Raman T, Subbaraya U (2023) Genome-wide characterization of 2OGD superfamily for mining of susceptibility factors responding to various biotic stresses in Musa spp. Physiol Mol Biol Plants 29(9):1319–1338. https://doi.org/10.1007/s12298-023-01380-y
Chelliah A, Arumugam C, Suthanthiram B, Raman T, Subbaraya U (2023) Genome-wide identification, characterization, and evolutionary analysis of NBS genes and their association with disease resistance in Musa spp. Funct Integr Genom 23(1):7. https://doi.org/10.1007/s10142-022-00925-w
Chen G, Cui J, Wang L, Zhu Y, Lu Z, Jin B (2017) Genome-wide identification of circular RNAs in Arabidopsis thaliana. Front Plant Sci 8:1678. https://doi.org/10.3389/fpls201701678
Chen L, Zhang P, Fan Y, Lu Q, Li Q, Yan J, Li L (2018) Circular RNAs mediated by transposons are associated with transcriptomic and phenotypic variation in maize. New Phytol 217(3):1292–1306. https://doi.org/10.1111/nph14901
Chen S, Zhou Y, Chen Y, Gu J (2018b) Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34(17):i884–i890. https://doi.org/10.1093/bioinformatics/bty560
Chidambara B, Elangovan D, Avverahally S, Reddy K, Kundapura R (2022) Identification of circular RNAs in resistant tomato genotype in response to ToLCBaV infection. J Hortic Sci 17(2):496–504. https://doi.org/10.24154/jhsv17i21520
Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res 39:W155–W159. https://doi.org/10.1093/nar/gkr319
Dai X, Zhuang Z, Zhao PX (2018) psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res 46(W1):W49–W54. https://doi.org/10.1093/nar/gky316
Dale J, James A, Paul JY, Khanna H, Smith M, Peraza-Echeverria S, Garcia-Bastidas F, Kema G, Waterhouse P, Mengersen K, Harding R (2017) Transgenic Cavendish bananas with resistance to Fusarium wilt tropical race 4. Nat Commun 8(1):1496. https://doi.org/10.1038/s41467-017-01670-6
Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, Li H (2021) Twelve years of SAMtools and BCFtools. GigaScience 10(2):giab008. https://doi.org/10.1093/gigascience/giab008
Darbani B, Noeparvar S, Borg S (2016) Identification of circular RNAs from the parental genes involved in multiple aspects of cellular metabolism in barley. Front Plant Sci 7:185329. https://doi.org/10.3389/fpls201600776
de Torres-Zabala M, Truman W, Bennett MH, Lafforgue G, Mansfield JW, Rodriguez Egea P, Bögre L, Grant M (2007) Pseudomonas syringae pv tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J 26(5):1434–1443. https://doi.org/10.1038/sjemboj7601575
Dong S, Wang Y (2016) Nudix Effectors: a common weapon in the arsenal of plant pathogens. PLoS Pathog 12(e10):05704. https://doi.org/10.1371/journalppat10.05704
Dou Y, Li S, Yang W, Liu K, Du Q, Ren G, Zhang C (2017) Genome-wide discovery of circular RNAs in the leaf and seedling tissues of Arabidopsis thaliana. Curr Genom 18(4):360–365. https://doi.org/10.2174/1389202918666170307161124
Droc G, Martin G, Guignon V et al (2022) The banana genome hub: a community database for genomics in the Musaceae. Horticult Res 9:uhac221. https://doi.org/10.1093/hr/uhac221
Fahlgren N (2010) Carrington JC (2010.) miRNA target prediction in plants. In: Meyers BC, Green PJ (eds) Plant MicroRNAs: Methods and Protocols, Humana Press: Totowa. NJ, USA, pp 51–57
Fan J, QuanW Li GB, Hu XH, Wang Q, Wang H, Li X, Luo X, Feng Q, Hu Z, Feng H, Pu M, Zhao J, Huang Y, Li Y, Zhang Y, Wang WM (2020) circRNAs are involved in the rice-Magnaporthe oryzae interaction. Plant Physiol 182(1):272–286. https://doi.org/10.1104/pp1900716
Fei S, Czislowski E, Fletcher S, Peters J, Batley J, Aitken E, Mitter N (2019) Small RNA profiling of Cavendish banana roots inoculated with Fusarium oxysporum f sp cubense race 1 and tropical race 4. Phytopathol Res 1:1–12. https://doi.org/10.1186/s42483-019-0029-3
Fourie G, Steenkamp ET, Ploetz RC, Gordon TR, Viljoen A (2011) Current status of the taxonomic position of Fusarium oxysporum f sp cubense within the Fusarium oxysporum complex. Infect Genet Evol 11(3):533–542. https://doi.org/10.1016/jmeegid20110.10.12
Gao Y, Wang J, Zheng Y, Zhang J, Chen S, Zhao F (2016) Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat Commun 7:12060. https://doi.org/10.1038/ncomms12060
Gao Y, Zhang J, Zhao F (2018) Circular RNA identification based on multiple seed matching. Brief Bioinform 19:803–810. https://doi.org/10.1093/bib/bbx014
Gao Z, Li J, Luo M, Li H, Chen Q, Wang L, Song S, Zhao L, Xu W, Zhang C, Wang S, Ma C (2019) Characterization and cloning of grape circular RNAs identified the cold resistance-related Vv-circATS1. Plant Physiol 180(2):966–985. https://doi.org/10.1104/pp1801331
García-Bastidas FA, Arango-Isaza R, Rodriguez-Cabal HA, Seidl MF, Cappadona G, Segura R, Salacinas M, Kema GH (2022) Induced resistance to Fusarium wilt of banana caused by Tropical Race 4 in Cavendish cv Grand Naine bananas after challenging with avirulent Fusarium spp. PLOS one 17(9):e0273335. https://doi.org/10.1371/journalpone0273335
Ge X, Xia Y (2008) The role of AtNUDT7, a nudix hydrolase, in the plant defense response. Plant Signal Behav 3(2):119–120. https://doi.org/10.4161/psb325019
Ge X, Li GJ, Wang SB, Zhu H, Zhu T, Wang X, Xia Y (2007) AtNUDT7, a negative regulator of basal immunity in Arabidopsis, modulates two distinct defense response pathways and is involved in maintaining redox homeostasis. Plant Physiol 145:204–215. https://doi.org/10.1104/pp10.710.3374
Ghorbani A, Izadpanah K, Peters JR, Dietzgen RG, Mitter N (2018) Detection and profiling of circular RNAs in uninfected and maize Iranian mosaic virus-infected maize. Plant Sci 274:402–409. https://doi.org/10.1016/jplantsci201806016
Guo Z, Kuang Z, Wang Y, Zhao Y, Tao Y, Cheng C, Yang J, Lu X, Hao C, Wang T, Cao X, Wei J, Li L, Yang X (2020) PmiREN: a comprehensive encyclopedia of plant miRNAs. Nucleic Acids Res 48(D1):D1114–D1121. https://doi.org/10.1093/nar/gkz894
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495(7441):384–388. https://doi.org/10.1038/nature11993
Hofius D, Li L, Hafrén A, Coll NS (2017) Autophagy as an emerging arena for plant–pathogen interactions. Curr Opin Plant Biol 38:117–123. https://doi.org/10.1016/jpbi201704017
Hong YH, Meng J, Zhang M, Luan YS (2020) Identification of tomato circular RNAs responsive to Phytophthora infestans. Gene 746:144652. https://doi.org/10.1016/jgene2020144652
Ishikawa K, Ogawa T, Hirosue E, Nakayama Y, Harada K, Fukusaki E, Yoshimura K, Shigeoka S (2009) Modulation of the poly(ADP-ribosyl)ation reaction via the Arabidopsis ADP-ribose/NADH Pyrophosphohydrolase, AtNUDX7, is involved in the response to oxidative stress. Plant Physiol 151:741–754. https://doi.org/10.1104/pp10.9140442
Ishikawa K, Yoshimura K, Ogawa T, Shigeoka S (2010) Distinct Regulation of Arabidopsis ADP-Ribose/NADH Pyrophosphohydrolases, Atnudx6 And 7, In Biotic and Abiotic Stress Responses. Plant Signal Behav 5:839–841. https://doi.org/10.4161/psb5711820
Jambunathan N, Mahalingam R (2006) Analysis of Arabidopsis growth factor gene 1 (AtGFG1) encoding a nudix hydrolase during oxidative signalling. Planta 224:1–11. https://doi.org/10.1007/s00425-005-0183-y
Jambunathan N, Penaganti A, Tang Y, Mahalingam R (2010) Modulation of redox homeostasis under suboptimal conditions by Arabidopsis nudix hydrolase 7. BMC Plant Biol 10:173. https://doi.org/10.1186/1471-2229-10-173
Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32(5):453–461. https://doi.org/10.1038/nbt2890
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y (2007) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36(suppl_1):D480–D484. https://doi.org/10.1093/nar/gkm882
Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC (2003) P1/HC-Pro, a viral suppressor of RNA silencing, interferes with arabidopsis development and miRNA. Function Dev Cell 4:205–217. https://doi.org/10.1016/s1534-5807(03)00025-x
Kim D, Paggi JM, Park C, Bennett C, Salzberg SL (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37(2019):907–915. https://doi.org/10.1038/s41587-019-0201-4
Koroban NV, Kudryavtseva AV, Krasnov GS, Sadritdinova AF, Fedorova MS, Snezhkina AV, Snezhkina AV, Bolsheva NL, Muravenko OV, Dmitriev AA, Melnikova NV (2016) The role of microRNA in abiotic stress response in plants. Mol Biol 50:337–343. https://doi.org/10.1002/tpg220350
Kwenda S, Motlolometsi TV, Birch PR, Moleleki LN (2016) RNA-seq profiling reveals defense responses in a tolerant potato cultivar to stem infection by Pectobacterium carotovorum ssp brasiliense. Front Plant Sci 7:238216. https://doi.org/10.3389/fpls201601905
Li CY, Deng GM, Yang J, Viljoen A, Jin Y, Kuang RB, Yi GJ (2012) Transcriptome profiling of resistant and susceptible Cavendish banana roots following inoculation with Fusarium oxysporum f sp cubense tropical race 4. BMC Genom 13:1–11. https://doi.org/10.1186/1471-2164-13-374
Li C, Shao J, Wang Y, Li W, Guo D, Yan B, Peng M (2013) Analysis of banana transcriptome and global gene expression profiles in banana roots in response to infection by race 1 and tropical race 4 of Fusarium oxysporum f sp cubense. BMC Genom 14:1–16. https://doi.org/10.1186/1471-2164-14-851
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Shan G (2015) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22(3):256–264. https://doi.org/10.1038/nsmb2959
Li Y, Zhao SL, Li JL, Hu XH, Wang H, Cao XL, Wang WM (2017) Osa-miR169 negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Front Plant Sci 8:234434. https://doi.org/10.3389/fpls201700002
Li Y, Jeyakumar JMJ, Feng Q, Zhao ZX, Fan J, Khaskheli MI, Wang WM (2019) The roles of rice microRNAs in rice-Magnaporthe oryzae interaction. Phytopathol Res 1(1):1–12. https://doi.org/10.1186/s42483-019-0040-8
Li M, Yang Z, Chang C (2022) Susceptibility is new resistance: wheat susceptibility genes and exploitation in resistance breeding. Agriculture 12(9):1419. https://doi.org/10.3390/agriculture12091419
Lin F, Zhao GA, Chen ZG, Wang XH, Lü FH, Zhang YC, Cai RY, Liang WQ, Li JH, Li M, Zhang GH, Yang YM (2018) Network correlation of circRNA-miRNA and the possible regulatory mechanism in acute myocardial infarction. Zhonghua Yi XueZaZhi 11:851–854. https://doi.org/10.3760/cmajissn0376-24912018110.12
Liu H, Nwafor CC, Piao Y, Li X, Zhan Z, Piao Z (2022) Identification and characterization of circular RNAs in Brassica rapa in response to Plasmodiophora brassicae. Int J Mol Sci 23:5369. https://doi.org/10.3390/ijms2310.5369
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion forRNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Low YC, Lawton MA, Di R (2020) Validation of barley 2OGO gene as a functional orthologue of Arabidopsis DMR6 gene in Fusarium head blight susceptibility. Sci Rep 10(1):9935. https://doi.org/10.1038/s41598-020-67006-5
Lu S, Sun YH, Amerson H, Chiang VL (2007) MicroRNAs in loblolly pine (Pinustaeda L) and their association with fusiform rust gall development. Plant J 51(6):10.77-10.98. https://doi.org/10.1111/j1365-313X200703208x
Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H, Han B (2015) Transcriptome-wide investigation of circular RNAs in rice. RNA 21(12):2076–2087. https://doi.org/10.1261/rna052282115
Luan Y, Cui J, Zhai J, Li J, Han L, Meng J (2015) High-throughput sequencing reveals differential expression of miRNAs in tomato inoculated with Phytophthora infestans. Planta 241:1405–1416. https://doi.org/10.1007/s00425-015-2267-7
Lv L, Yu K, Lü H, Zhang X, Liu X, Sun C, Zhang D (2020) Transcriptome-wide identification of novel circular RNAs in soybean in response to low-phosphorus stress. PloS one 15(1):e0227243. https://doi.org/10.1371/journalpone0227243
Mäkinen V, Välimäki N, Sirén J (2014) Indexing graphs for path queries with applications in genome researchIEEE/ACM Trans. Comput Biol Bioinform 11:375–388
Mammarella ND, Cheng Z, Fu ZQ, Daudi A, Bolwell GP, Dong X, Ausubel FM (2015) Apoplastic peroxidases are required for salicylic acid-mediated defense against Pseudomonas syringae. Phytochem 112:110–121. https://doi.org/10.1016/jphytochem201407010
Mao X, Cai T, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793. https://doi.org/10.1093/bioinformatics/bti430
McGinnis S, Madden TL (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 32(Web Server issue):W20–W25. https://doi.org/10.1093/nar/gkh435
Mostert D, Molina AB, Daniells J, Fourie G, Hermanto C, Chao CP, Viljoen A (2017) The distribution and host range of the banana Fusarium wilt fungus, Fusarium oxysporum f sp cubense. Asia. PLoS One 12(7):e0181630. https://doi.org/10.1371/journalpone0181630
Ouyang S, Park G, Atamian HS, Han CS, Stajich JE, Kaloshian I, Borkovich KA (2014) MicroRNAs suppress NB domain genes in tomato that confer resistance to Fusarium oxysporum. PLoS Pathog 10(10):e1004464. https://doi.org/10.1371/journalppat10.04464
Padmanabhan C, Zhang X, Jin H (2009) Host small RNAs are big contributors to plant innate immunity. Curr Opin Plant Biol 12(4):465–472. https://doi.org/10.1016/jpbi200906005
Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, Shenzis S, Samson M, Dittmar G, Landthaler M, Chekulaeva M, Rajewsky N, Kadener N (2017) Translation of CircRNAs. Mol Cell 66:9–217. https://doi.org/10.1016/jmolcel201702021
Pan T, Sun X, Liu Y, Li H, Deng G, Lin H, Wang S (2018) Heat stress alters genome-wide profiles of circular RNAs in Arabidopsis. Plant Mol Biol 96(3):217–29. https://doi.org/10.1007/s1110.3-017-0684-7
Pan J, Zhang X, Zhang Y, Yan B, Dai K, Zhu M, Gong C (2021) Grass carp reovirus encoding circular RNAs with antiviral activity. Aquac 533:736135. https://doi.org/10.1016/jaquaculture2020736135
Park JH, Shin C (2015) The role of plant small RNAs in NB-LRR regulation. Brief Funct Genom 14(4):268–274. https://doi.org/10.1093/bfgp/elv006
Peraza-Echeverria S, Dale JL, Harding RM, Smith MK, Collet C (2008) Characterization of disease resistance gene candidates of the nucleotide binding site (NBS) type from banana and correlation of a transcriptional polymorphism with resistance to Fusarium oxysporum fsp cubense race 4. Mol Breed 22(4):565–579. https://doi.org/10.1007/s110.32-008-9199-x
Peraza-Echeverria S, Dale JL, Harding RM, Collet C (2009) Molecular cloning and in silico analysis of potential Fusarium resistance genes in banana. Mol Plant Breed 23:431–443. https://doi.org/10.1007/s110.32-008-9247-6
Ploetz RC (2005) Panama disease: an old nemesis rears its ugly head: Part 1 The beginnings of the banana export trades. PHP 6(1):18. https://doi.org/10.1094/PHP-2005-1221-01-RV
Ploetz RC, Correll JC (1988) Vegetative compatibility among races of Fusarium oxysporum f sp cubense. Plant Dis 72:325–328
Ploetz RC, Pegg KG (2000) Fusarium wilt. In: Jones DR (ed) Disease of Banana, Handbook of diseases of banana, abacá and enset. CABI Publishing, Wallingford, UK, pp 143–159
Sablok G, Zhao H, Sun X (2016) Plant circular RNAs (circRNAs): transcriptional regulation beyond miRNAs in plants. Mol Plant 9(2):192–194. https://doi.org/10.1016/jmolp201512021
Seitz H (2009) Redefining microRNA targets. Curr Biol 19(10):870–873. https://doi.org/10.1016/jcub200903059
ShannonP MA, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–504. https://doi.org/10.1101/gr1239303
Song S, Chen X, Huang D, XuY ZH, Hu X, Wang W (2016) Identification of miRNAs differentially expressed in Fusarium wilt-resistant and susceptible banana varieties. S Afr J Bot 10(6):244–249. https://doi.org/10.1016/jsajb201606007
Song S, Xu Y, Huang D, Ashraf MA, Li J, Hu W, Xie J (2018) Identification and characterization of miRNA169 family members in banana (Musa acuminata L) that respond to fusarium oxysporum f sp cubense infection in banana cultivars. Peer J 6:e6209. https://doi.org/10.7717/peerj6209
Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A (2015) Exon circularization requires canonical splice signals. Cell Rep 10:103–111. https://doi.org/10.1016/jcelrep201412002
Sun K, Wolters AA, Vossen JH, Rouwet ME, Loonen AE, Jacobsen E, Visser R, Bai Y (2016) Silencing of six susceptibility genes results in potato late blight resistance. Transgenic Res 25(5):731–742. https://doi.org/10.1007/s11248-016-9964-2
Sun Y, Fan M, He Y (2019) Transcriptome analysis of watermelon leaves reveals candidate genes responsive to cucumber green mottle mosaic virus infection. Int J Mol Sci 20(3):610. https://doi.org/10.3390/ijms20030610
Sun Y, Zhang H, Fan M, He Y, Guo P (2020) Genome-wide identification of long non-coding RNAs and circular RNAs reveal their ceRNA networks in response to cucumber green mottle mosaic virus infection in watermelon. Arch Virol 165(5):1177–1190. https://doi.org/10.1007/s00705-020-04589-4
Sun J, Dong Y, Wang C, Xiao S, Jiao Z, Gao C (2021) Identification and characterization of melon circular RNAs involved in powdery mildew responses through comparative transcriptome analysis. PeerJ 9:e11216. https://doi.org/10.7717/peerj11216
Swarupa V, Ravishankar KV, Rekha A (2013) Characterization of tolerance to Fusarium oxysporum f sp, cubense infection in banana using suppression subtractive hybridization and gene expression analysis. Physiol Mol Plant Pathol 83:1–7. https://doi.org/10.1016/jpmpp201302003
Tong W, Yu J, Hou Y, Li F, Zhou Q, Wei C, Bennetzen JL (2018) Circular RNA architecture and differentiation during leaf bud to young leaf development in tea (Camellia sinensis). Planta 248:1417–1429. https://doi.org/10.1007/s00425-018-2983-x
Törönen P, Holm L (2022) PANNZER-A practical tool for protein function prediction. Protein Sci 31(1):118–128. https://doi.org/10.1002/pro4193
vanDamme M, Huibers RP, Elberse J, Van den Ackerveken G (2008) Arabidopsis DMR6 encodes a putative 2OG-Fe(II) oxygenase that is defence-associated but required for susceptibility to downy mildew. Plant J 54:785–793. https://doi.org/10.1111/j1365-313X200803427x
vanSchie CC, Takken FL (2014) Susceptibility genes 10.1: how to be a good host. Annu Rev Phytopathol 52:551–581. https://doi.org/10.1146/annurev-phyto-10.2313-045854
Vicens Q, Westhof E (2014) Biogenesis of circular RNAs. Cell 159(1):13–14. https://doi.org/10.1016/jcell201409005
Wang Y, Wang Z (2015) Efficient backsplicing produces translatable circular mRNAs. RNA 21(2):172–179. https://doi.org/10.1261/rna048272114
Wang Z, Zhang J, Jia C, Liu J, Li Y, Yin X, Jin Z (2012) De novo characterization of the banana root transcriptome and analysis of gene expression under Fusarium oxysporum f sp cubense tropical race 4 infection. BMC Genom 13:1–9. https://doi.org/10.1186/1471-2164-13-650
Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Salzman J (2014) Circular RNA is expressed across the eukaryotic tree of life. PloS one 9(3):e90859. https://doi.org/10.1371/journalpone0090859
Wang Y, Wang Q, Gao L, Zhu B, Luo Y, Deng Z, Zuo J (2017) Integrative analysis of circRNAs acting as ceRNAs involved in ethylene pathway in tomato. Physiol Plant 161(3):311–321. https://doi.org/10.1111/ppl12600
Wang Y, Yang M, Wei S, Qin F, Zhao H, Suo B (2017) Identification of circular RNAs and their targets in leaves of Triticum aestivum L under dehydration stress. Front Plant Sci 7:2024. https://doi.org/10.3389/fpls201602024
Wang J, Yang Y, Jin L, Ling X, LiuT CT, Zhang B (2018) Re-analysis of long non-coding RNAs and prediction of circRNAs reveal their novel roles in susceptible tomato following TYLCV infection. BMC Plant Boil 18:1–16. https://doi.org/10.1186/s12870-018-1332-3
Wang W, Wang J, Wei Q, Li B, Zhong X, Hu T, Bao C (2019) Transcriptome-wide identification and characterization of circular RNAs in leaves of Chinese cabbage (Brassica rapa L ssp pekinensis) in response to calcium deficiency-induced tip-burn. Sci Rep 9(1):14544. https://doi.org/10.1038/s41598-019-51190-0
Wu HJ, Wang ZM, Wang M, Wang XJ (2013) Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol 161(4):1875–1884. https://doi.org/10.1104/pp113215962
Xiang L, Cai C, Cheng J, Wang L, Wu C, Shi Y, Cai Y (2018) Identification of circularRNAs and their targets in Gossypium under Verticillium wilt stress based on RNA-seq. Peer J 6:e4500. https://doi.org/10.7717/peerj4500
Xin M, Wang Y, Yao Y, Xie C, Peng H, Ni Z, Sun Q (2010) Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L). BMC Plant Biol 10:123. https://doi.org/10.1186/1471-2229-10-123
Yang L, Mu X, Liu C, Cai J, Shi K, Zhu W, Yang Q (2015) Over-expression of potato miR482e enhanced plant sensitivity to Verticilliumdahliae infection. J Integr Plant Biol 57:1078–1088. https://doi.org/10.1111/jipb12348
Yang Q, Huai B, Lu Y, Cai K, Guo J, Zhu X, Kang Z, Guo J (2020) A stripe rust effector Pst18363 targets and stabilises TaNUDX23 that promotes stripe rust disease. New Phytol 225:880–895. https://doi.org/10.1111/nph16199
Yang X, Zhang L, Yang Y, Schmid M, Wang Y (2021) miRNA mediated regulation and interaction between plants and pathogens. Int J Mol Sci 22(6):2913. https://doi.org/10.3390/ijms22062913
Ye CY, Chen L, Liu C, Zhu QH, Fan L (2015) Widespread noncoding circular RNA s in plants. New Phytol 208(1):88–95. https://doi.org/10.1111/nph13585
Ye CY, Zhang X, Chu Q, Liu C, Yu Y, Jiang W, Guo L (2017) Full-length sequence assembly reveals circular RNAs with diverse non-GT/AG splicing signals in rice. RNA Biol 14(8):1055–1063. https://doi.org/10.1080/1547628620161245268
Yin J, Liu M, Ma D, Wu J, Li S, Zhu Y, Han B (2018) Identification of circular RNAs and their targets during tomato fruit ripening. Postharvest Biol Tec 136:90–98. https://doi.org/10.1016/jpostharvbio201710.013
Yoshimura K, Shigeoka S (2015) Versatile physiological functions of the Nudix hydrolase family in Arabidopsis. Biosci Biotechnol Biochem 79(3):354–66. https://doi.org/10.1080/091684512014987207
Zhang W, Gao S, Zhou X, Chellappan P, Chen Z, Zhou X, Zhang X, Fromuth N, Coutino G, Coffey M, Jin H (2011) Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol Biol 75(93–10):5. https://doi.org/10.1007/s1110.3-010-9710.-8
Zhang K, Halitschke R, Yin C, Liu CJ, Gan SS (2013) Salicylic acid 3-hydroxylase regulates Arabidopsis leaf longevity by mediating salicylic acid catabolism. Proc Natl Acad Sci USA 110:14807–14812. https://doi.org/10.1073/pnas1302702110
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Chen LL (2013) Circular intronic long noncoding RNAs. Mol Cell 51(6):792–806. https://doi.org/10.1016/jmolcel201308017
Zhang T, Zhao YL, Zhao JH, Wang S, Jin Y, Chen ZQ, Fang YY, Hua CL, Ding SW, Guo HS (2016) Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat Plants 2(10):1–6. https://doi.org/10.1038/nplants2016153
Zhang QL, Ji XY, Li HW, Guo J, Wang F, Deng XY, Lin LB (2019) Identification of circular RNAs and their altered expression under poly (I: C) challenge in key antiviral immune pathways in amphioxus Fish Shellfish. Immunol 86:1053–1057. https://doi.org/10.1016/jfsi201812061
Zhao W, Cheng Y, Zhang C, You Q, Shen X, Guo W, Jiao Y (2017) Genome-wide identification and characterization of circular RNAs by high throughput sequencing in soybean. Sci Rep-UK 7(1):5636. https://doi.org/10.1038/s41598-017-05922-9
Zhou R, Zhu Y, Zhao J, Fang Z, Wang S, Yin J, Chu Z, Ma D (2017) Transcriptome-wide identification and characterization of potato circular RNAs in response to Pectobacterium carotovorum subspecies brasiliense infection. Int J Mol Sci 19(1):71. https://doi.org/10.3390/ijms19010.071
Zhu QH, Fan L, Liu Y, Xu H, Llewellyn D, Wilson I (2013) miR482 regulation of NBS-LRR defense genes during fungal pathogen infection in cotton. PloS one 8(12):e84390. https://doi.org/10.1371/journalpone0084390
Zhu CL, Sha X, Wang Y, Li J, Zhang MY, Guo ZY, Sun S, He JD (2019) Circular RNA hsa_circ_0007142 is upregulated and targets miR-103a-2–5p in colorectal cancer. J Oncol 2019:9836819. https://doi.org/10.1155/2019/9836819
Zuo J, Wang Q, Zhu B, Luo Y, Gao L (2016) Deciphering the roles of circRNAs on chilling injury in tomato. Biochem Bioph Res Co 479(2):132–138. https://doi.org/10.1016/jbbrc201607032
Author Information
Crop Improvement Division, ICAR-National Research Centre for Banana, Tiruchirappalli, India