Whole genome sequencing, assembly and annotation of Curvularia geniculata strain SKG23 isolated from leaf spot disease of mentha (Mentha arvensis L.)
*Article not assigned to an issue yet
Research Articles | Published: 09 October, 2025
First Page: 0
Last Page: 0
Views: 112
Keywords: Fungi, Genome, Sequence, Assembly, Annotation
Abstract
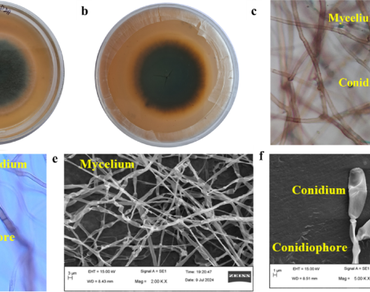
A strain of fungus was isolated and purified from a diseased leaf of Mentha (M. arvensis L.), and it was molecularly identified as Curvularia geniculata strain SKG23. The whole genome sequence of this fungal strain was done, and it showed that its genome size was 38.36 Mb. The current genome assembly of the C. geniculata strain SKG23 consisted of 4655 scaffolds with an N50 length of 196,991 bp and an N90 length of 4088 bp. GC (guanine-cytosine) content and repetitive element of this strain were 51.56 and 0.68%, respectively. In this strain, 17,271 predicted protein-coding genes, or total genes, were present, and CAZymes (carbohydrate-active enzymes) were 287. Number of non-CAZyme transmembrane transporter protein-coding genes that were annotated was 979 genes, which was the highest among fungi so far reported, and among them MFS (major facilitator superfamily) transporters were the highest in number (350), followed by amino acid (156), peptide (142), sugar transporters (79), ABC (ATP-binding cassette) transporters (109), etc., and non-transporter genes were also annotated in the genome of this strain. This draft genome data may be a good source for further research on fungi.
References
Acharya MC (2024) Curvularia verruculosa strain: KHW-7 genome sequencing and assembly. PRJNA1023514, NCBI, Baltimore, USA
Ali SS, Asman A, Shao J et al (2020) Genome and transcriptome analysis of the latent pathogen Lasiodiplodia theobromae, an emerging threat to the cacao industry. Genome 63:37–52. https://doi.org/10.1139/gen-2019-0112
Alonge M, Lebeigle L, Kirsche M et al (2021) Automated assembly scaffolding elevates a new tomato system for high-throughput genome editing. https://doi.org/10.1101/2021.11.18.469135. BioRxiv.
Anonymous (2021) Curvularia sp. ZM96 isolated from saline soils on the Tibetan Plateau, Shaanxi university of science and technology. PRJNA775668, European Nucleotide Archive (ENA)
Anonymous (2018) Curvularia geniculata strain:W3 genome sequencing and assembly. PRJNA432274, Universidade federal de Mato Grosso- UFMT, PRJNA432274. NCBI, USA
Bailey BA, Melnick RL, Strem MD et al (2014) Differential gene expression by M oniliophthora roreri while overcoming cacao tolerance in the field. Mol Plant Pathol 15:711–729. https://doi.org/10.1111/mpp.12134
Beseli A, Amnuaykanjanasin A, Herrero S et al (2015) Membrane transporters in self resistance of Cercospora nicotianae to the photoactivated toxin Cercosporin. Curr Genet 61:601–620. https://doi.org/10.1007/s00294-015-0486-x
Borodovsky M, Lomsadze A (2011) Eukaryotic gene prediction using GeneMark.hmm-E and GeneMark-ES. Curr Protoc Bioinf. 1(1):4–5. https://doi.org/10.1002/0471250953.bi0406s35. 4.6.1–4.6.10
Cao Y, Cai Q, Li C et al (2023) Genome sequence resource of curvularia clavata causing leaf spot disease on tobacco by Oxford Nanopore PromethION. Plant Dis 107:1916–1919. https://doi.org/10.1094/PDIS-09-22-2283-A
Chethana KT, Li X, Zhang W et al (2016) Trail of decryption of molecular research on botryosphaeriaceae in woody plants. Phytopathol Mediterr 147–171. https://doi.org/10.14601/Phytopathol_Mediterr-16230
Chomnunti P, Schoch CL, Aguirre-Hudson B et al (2011) Capnodiaceae. Fungal Divers 51:103–134.
Crous PW, Cowan DA, Maggs-Kölling G et al (2021) Fungal planet description sheets. Pers: Mol Phylogeny Evol Fungi 46:1182–1283.
da Cunha KC, Sutton DA, Fothergill AW et al (2013) In vitro antifungal susceptibility and molecular identity of 99 clinical isolates of the opportunistic fungal genus Curvularia. Diagn Microbiol Infect Dis 76:168–174.
Davet P, Rouxel F (2000) Detection and isolation of soil fungi. Science Publishers, USA, pp 188.
Dilokpimol A, Mäkelä MR, Aguilar-Pontes MV et al (2016) Diversity of fungal feruloyl esterases: updated phylogenetic classification, properties, and industrial applications. Biotechnol Biofuels 9:1–18. https://doi.org/10.1186/s13068-016-0651-6
Domsch KH, Gams W, Anderson TH (1980) Compendium of soil fungi, vol 1. Academic Press (London) Ltd. London.
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem bull. 19(1):11–15
El-Gebali S, Mistry J, Bateman A et al (2019) The Pfam protein families database in 2019. Nucleic Acids Res 47:D427–D432. https://doi.org/10.1093/nar/gky995
Emms DM, Kelly S (2019) OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol 20:1–14. https://doi.org/10.1186/s13059-019-1832-y
Ferdinandez HS, Manamgoda DS, Udayanga D et al (2024) Molecular phylogeny and morphology reveal two new graminicolous species, Curvularia aurantia sp. nov. and C. vidyodayana sp. nov. with new records of Curvularia spp. from Sri Lanka. Fung Syst Evol 12:219–246. https://doi.org/10.3114/fuse.2023.12.11
Galagan JE (2014) Genomic insights into tuberculosis. Nat Rev Genet 15:307–320. https://doi.org/10.1038/nrg3664
Galagan JE, Calvo SE, Borkovich KA et al (2003) The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859–868. https://doi.org/10.1038/nature01554
Gao S, Li Y, Gao J et al (2014) Genome sequence and virulence variation-related transcriptome profiles of Curvularia lunata, an important maize pathogenic fungus. BMC Genomics 15:1–18. https://doi.org/10.1186/1471-2164-15-627
Ghosh SK, Pal S (2021) De-polymerization of LDPE plastic by Penicillium simplicissimum isolated from municipality garbage plastic and identified by ITSs locus of rDNA. Vegetos 34:57–67. https://doi.org/10.1007/s42535-020-00176-9
Ghosh SK, Pal S, Banerjee S (2022) Identification and pathogenicity of Alternaria alternata causing leaf blight of Bacopa monnieri (L.) Wettst. and its biocontrol by Trichoderma species in agrifields–an ecofriendly approach. J Appl Res Med Aromat Plants 31:100406. https://doi.org/10.1016/j.jarmap.2022.100406Get
Ghosh SK, Pal S, Ray S (2013) Study of microbes having potentiality for biodegradation of plastics. Environ Sci Pollut Res 20:4339–4355. https://doi.org/10.1007/s11356-013-1706-x
Goffeau A, Barrell BG, Bussey H et al (1996) Life with 6000 genes. Science 274:546–567. https://doi.org/10.1126/science.274.5287.546
Han WB, Lu YH, Zhang AH, Zhang GF, Mei YN, Jiang N, Lei X, Song YC, Ng SW, Tan RX (2014) Curvulamine, a new antibacterial alkaloid incorporating two undescribed units from a Curvularia species. Org Lett 16(20):5366–5369. https://doi.org/10.1021/ol502572g
Handayani D, Putri RA, Ismed F et al (2020) Bioactive metabolite from marine sponge-derived fungus Cochliobolus geniculatus WR12. Rasayan J Chem 13:417–422. https://doi.org/10.31788/RJC.2020.1315517
Huerta-Cepas J, Forslund K, Coelho LP et al (2017) Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol 34:2115–2122. https://doi.org/10.1093/molbev/msx148
Kaboré KH, Kassankogno AI, Adreit H et al (2023) Whole-genome sequences of bipolaris bicolor, curvularia hawaiiensis, curvularia spicifera, and exserohilum rostratum isolated from rice in Burkina Faso, France, Mali, and Pakistan. Microbiol Resour Announc 12:e00134–e00123.
Kee YJ, Zakaria L, Mohd MH (2020) Curvularia asianensis and Curvularia Eragrostidis associated with leaf spot of Sansevieria trifasciata in Malaysia. J Phytopathol 168:290–296.
Lo Presti L, Lanver D, Schweizer G et al (2015) Fungal effectors and plant susceptibility. Annu Rev Plant Biol 66:513–545. https://doi.org/10.1146/annurev-arplant-043014-114623
Luo R, Liu B, Xie Y et al (2015) Erratum: SOAPdenovo2: an empirically improved memory-efficient short-read de Novo assembler. Gigascience 4:1–1.
Madrid H, Da Cunha KC, Gené J et al (2014) Novel Curvularia species from clinical specimens. Pers: Mol Phylogeny Evol Fungi 33:48–60. https://doi.org/10.3767/003158514x683538
Manamgoda DS, Cai L, McKenzie EH et al (2012) A phylogenetic and taxonomic re-evaluation of the Bipolaris-Cochliobolus-Curvularia complex. Fungal Divers 56:131–144. https://doi.org/10.1007/s13225-012-0189-2
Manamgoda DS, Rossman A, Castlebury LA et al (2015) Article PHYTOTAXA. Phytotaxa 212:175–198. https://doi.org/10.11646/phytotaxa.212.3.1
Meinhardt LW, Costa GGL, Thomazella DP et al (2014) Genome and secretome analysis of the hemibiotrophic fungal pathogen, Moniliophthora roreri, which causes frosty pod rot disease of cacao: mechanisms of the biotrophic and necrotrophic phases. BMC Genomics 15:1–25. https://doi.org/10.1186/1471-2164-15-164
Morales-Cruz A, Amrine KC, Blanco-Ulate B et al (2015) Distinctive expansion of gene families associated with plant cell wall degradation, secondary metabolism, and nutrient uptake in the genomes of grapevine trunk pathogens. BMC Genomics 16:1–22. https://doi.org/10.1186/s12864-015-1624-z
Nagamani A, Kunwar IK, Manoharachary C (2006) Handbook of soil fungi. IK international. India.
Paolinelli-Alfonso M, Villalobos-Escobedo JM, Rolshausen P et al (2016) Global transcriptional analysis suggests Lasiodiplodia theobromae pathogenicity factors involved in modulation of grapevine defensive response. BMC Genomics 17:1–20.
Perlin MH, Andrews J, San Toh S (2014) Essential letters in the fungal alphabet: ABC and MFS transporters and their roles in survival and pathogenicity. Adv Genet 85:201–253.
Quach NT, Ngo CC, Nguyen TH et al (2022) Genome-wide comparison deciphers lifestyle adaptation and glass biodeterioration property of Curvularia Eragrostidis C52. Sci Rep 12:11411. https://doi.org/10.1038/s41598-022-15334-z
Schoch CL, Ciufo S, Domrachev M et al (2020) NCBI taxonomy: a comprehensive update on curation, resources and tools. Database.https://doi.org/10.1093/database/baaa062
Simão FA, Waterhouse RM, Ioannidis P et al (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinform 31:3210–3212. https://doi.org/10.1093/bioinformatics/btv351
Sivanesan A (1987) Graminicolous species of Bipolaris, Curvularia, Drechslera, Exserohilum and their teleomorphs. C.A.B. International. UK.
Smit AFA, Hubley R, Green P (2013–2015) RepeatMasker Open-4.0. http://www.repeatmasker.org
van den Brink HM, van Gorcom RF, van den Hondel CA et al (1998) Cytochrome P450 enzyme systems in fungi. Fungal Genet Biol 23:1–17. https://doi.org/10.1006/fgbi.1997.1021
Vankuyk PA, Diderich JA, MacCABE AP et al (2004) Aspergillus Niger MstA encodes a high-affinity sugar/H + symporter which is regulated in response to extracellular pH. Biochem J 379:375–383. https://doi.org/10.1042/bj20030624
Wang Y, Pan XJ, Zhang Q et al (2018) First report of Curvularia asianensis causing leaf blotch of Epipremnum pinnatum in Guangxi autonomous region of China. Plant Dis 102:1854. https://doi.org/10.1094/PDIS-01-18-0145-PDN
Wingett SW, Andrews S (2018) FastQ screen: a tool for multi-genome mapping and quality control. Natl Cent Biotechnol Inf. 7:1338. .
Wood V, Gwilliam R, Rajandream MA et al (2002) The genome sequence of Schizosaccharomyces Pombe. Nature 415:871–880.
Xue M, Yang J, Li Z et al (2012) Comparative analysis of the genomes of two field isolates of the rice blast fungus Magnaporthe oryzae. PLoS Genet 8:e1002869.
Yan JY, Zhao WS, Chen Z et al (2018) Comparative genome and transcriptome analyses reveal adaptations to opportunistic infections in Woody plant degrading pathogens of botryosphaeriaceae. DNA Res 25:87–102.
Ye C, Ma ZS, Cannon CH et al (2012) Exploiting sparseness in de Novo genome assembly. BMC Bioinformatics 13:1–8. https://doi.org/10.1186/1471-2105-13-s6-s1
Zhang H, Yohe T, Huang L et al (2018) dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46:W95–W101.https://doi.org/10.1093/nar/gky418
Zhang Y, Zhang K, Fang A et al (2014) Specific adaptation of Ustilaginoidea virens in occupying host florets revealed by comparative and functional genomics. Nat Commun 5:3849. https://doi.org/10.1038/ncomms4849
Zhang Y, Zhang Z, Zhang X et al (2012) CDR4 is the major contributor to Azole resistance among four Pdr5p-like ABC transporters in Neurospora crassa. Fungal Biol 116:848–854. https://doi.org/10.1016/j.funbio.2012.05.002
Author Information
Molecular Mycopathology Lab, Biocontrol Unit and Cancer Research Unit, Ramakrishna Mission Vivekananda Centenary College (Autonomous), Rahara, India