Whole proteome analysis of xero-halophyte Atriplex under salinity
Research Articles | Published: 23 August, 2022
First Page: 805
Last Page: 815
Views: 3547
Keywords: Atriplex , Comparative proteomics, Salt stress, Signaling, Xero-halophyte
Abstract
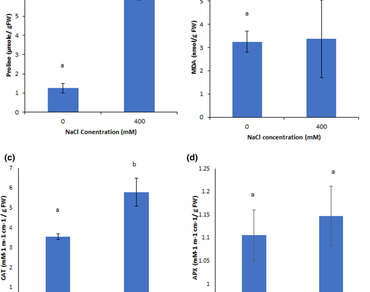
Salt stress is among the prime abiotic stresses that adversely affects crop yield worldwide. Halophytes are able to tolerate high salt concentrations, but a little information is available about their salt-adaptation mechanism. In the present study, we have investigated the molecular mechanism of salinity tolerance in Atriplex griffithii, an important xero-halophytic plant. Two-dimensional gel electrophoresis based comparative proteomics method was used to evaluate proteins’ expression in A. griffithii under control (0 mM NaCl) and saline (400 mM NaCl) conditions. Our results revealed that A. griffithii has strong potential to tolerate high concentration of salt, i.e., up to 400 mM. A total of 170 proteins were observed in both control and treatment gels, and 61 protein spots exhibited significant change of minimum twofold expression after imposing salt stress, with 11 novel salt-induced proteins. Maximum protein spots were having molecular weight and isoelectric point in a range of 14–110 kDa and 4–7, respectively. This study has revealed some important aspects of halophytic salt-stress responses at global proteome level. Our physiological data for proline content, lipid peroxidation and anti-oxidant enzymes activity also supported the salt-tolerant nature of this species. The differentially expressed proteins found in our study may be utilized to enhance abiotic stress tolerance in the sensitive agricultural crops through biotechnological techniques.
References
Barkla BJ, Vera-Estrella R, Hernandez-Coronado M, Pantoja O (2009) Quantitative proteomics of the tonoplast reveals a role for glycolytic enzymes in salt tolerance. Plant Cell 21:4044–4058
Dani V, Simon WJ, Duranti M, Croy RR (2005) Changes in the tobacco leaf apoplast proteome in response to salt stress. Proteomics 5(3):737–745
Flowers TJ, Colmer TD (2008) Salinity tolerance in halophytes. Tansley Rev-New Phytol 179:945–963
Glenn EP, Brown JJ (1998) Effects of soil salt levels on the growth and water use efficiency of Atriplex canescens (Chenopodiaceae) varieties in drying soil. Am J Bot 85:10–16
Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol 51:463–499
Hassine AB, Ghanem ME, Bouzid S, Lutts S (2008) An inland and a coastal population of the Mediterranean xero-halophyte species Atriplex halimus L. differ in their ability to accumulate proline and glycinebetaine in response to salinity and water stress. J Exp Bot 59(6):1315–1326
Iqbal MZ, Shafiq M, Athar M (2014) Adaptation in Atriplex griffithii and Prosopis juliflora plants in response to cement dust pollution. J Appl Sci Environ Manage 18(3):389–395
Jacoby RP, Millar AH, Taylor NL (2010) Wheat mitochondrial proteomes provide new links between antioxidant defense and plant salinity tolerance. J Proteome Res 9(12):6595–6604
Jha S (2018) Proteomics of Salinity Stress: Opportunities and Challenges. In: Ramakrishna A, Gill SS (eds) Metabolic Adaptations in Plants during Abiotic Stress. CRC Press, Taylor & Francis Group Boca Raton FL, pp 285–294
Jha S (2022) Proteome responses of pearl millet genotypes under salinity. Plant Gene 29:100347. https://doi.org/10.1016/j.plgene.2021.100347
Jha S, Agarwal S, Sanyal I, Amla DV (2012) Differential subcellular targeting of recombinant human α1-proteinase inhibitor influences yield, biological activity and in planta stability of the protein in transgenic tomato plants. Plant Sci 196:53–66
Jha S, Agarwal S, Sanyal I, Amla DV (2016) Single-step purification and characterization of a recombinant serine proteinase inhibitor from transgenic plants. Appl Biochem Biotechnol 179:220–236
Jha S (2019) Transgenic Approaches for Enhancement of Salinity Stress Tolerance in Plants. In: Singh SP et al (eds) Molecular Approaches in Plant Biology and Environmental Challenges. Springer Nature, Singapore, pp 265–322
Jha S, Maity S, Singh J, Chouhan C, Tak N, Ambatipudi K (2022) Integrated physiological and comparative proteomics analysis of contrasting genotypes of pearl millet reveals underlying salt-responsive mechanisms. Physiol Plant. https://doi.org/10.1111/ppl.13605
Jha S, Singh J, Chouhan C, Singh O, Srivastava RK (2022) Evaluation of multiple salinity tolerance indices for screening and comparative biochemical and molecular analysis of pearl millet [Pennisetum glaucum (L.) R .Br.] genotypes. J Plant Growth Regul. 41:1820–1834. https://doi.org/10.1007/s00344-021-10424-0
Jiang Y, Yang B, Harris NS, Deyholos MK (2007) Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J Exp Bot 58(13):3591–3607
Khan MA, Rizvi Y (1994) Effect of salinity, temperature, and growth regulators on the germination and early seedling growth of Atriplex griffithii var. stocksii. Can J Bot 72:475–479
Li Q, Yin H, Li D, Zhu H, Zhang Y, Zhu W (2007) Isolation and characterization of CMO promoter from Halophyte Sueada liaotungensis K. J Genet Genomics 34(4):355–361
Li W, Zhang C, Lu Q, Wen X, Lu C (2011) The combined effect of salt stress and heat shock on proteome profiling in Suaeda salsa. J Plant Physiol 168:1743–1752
Munns R (2002) Comparative physiology of salt and water stress. Plant Cell Environ 25:239–250
Munns R (2005) Genes and salt tolerance: Bringing them together. Tansley Rev New Phytol 167:645–663
Ndimba BK, Chivasa S, Simon WJ, Slabas AR (2005) Identification of Arabidopsis salt and osmotic stress responsive proteins using two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics 5:4185–4196
Panda A, Rangani J, Parida AK (2020) Comprehensive proteomic analysis revealing multifaceted regulatory network of the xero-halophyte Haloxylon salicornicum involved in salt tolerance. J Biotechnol 324:143–161
Pang Q, Chen S, Dai S, Chen Y, Wang Y, Yan X (2010) Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila. J Proteome Res 9:2584–2599
Sadder MT, Anwar F, Al-Doss AA (2013) Gene expression and physiological analysis of Atriplex halimus (L.) under salt stress. Aus J Crop Sci 7(1):112–118
Sangam S, Jayasree D, Janardham Reddy K, Chari PVB, Sreenivasulu N, Kavi Kishor PR (2005) Salt tolerance in plants-transgenic approaches. J Plant Biotechnol 7(1):1–15
Sengupta S, Majumder AL (2009) Insight into the salt tolerance factors of a wild halophytic rice, Porteresia. coarctata: A physiological and proteomic approach. Planta 229:911–929
Tada Y, Kashimura T (2009) Proteomic analysis of salt-responsive proteins in the mangrove plant. Bruguiera Gymnorhiza Plant Cell Physiol 50(3):439–446
Veeranagamallaiah G, Jyothsnakumari G, Thippeswamy M, Chandra Obul Reddy P, Surabhi G-K, Sriranganayakulu G, Mahesh Y, Rajasekhar B, Madhurarekha C, Sudhakar C (2008) Proteomic analysis of salt stress responses in foxtail millet (Setaria italica L. cv. Prasad) seedling. Plant Sci 175(5):631–641
Wang X, Fan P, Song H, Chen X, Li X, Li Y (2009) Comparative proteomic analysis of differentially expressed proteins in shoots of Salicornia europaea under different salinity. J Proteome Res 8:3331–3345
Author Information
Plant Functional Genomics Lab, Biotechnology Unit, Department of Botany (UGC-Centre of Advance Study), Jai Narain Vyas University, Jodhpur, India