Assessment of genetic diversity and phenetic relationships among some Bangladeshi cultivars of cucumber (Cucumis sativus L.) using isozyme and protein profiling
Habiba, Sarkar Dilruba, Haque Md. Enamul, Islam Md. Asadul, Mukherjee Ashutosh, Sikdar Biswanath
Research Articles | Published: 21 October, 2019
First Page: 494
Last Page: 507
Views: 4045
Keywords: Jaccard similarity, Isozyme banding pattern, SDS-PAGE, UPGMA, Zymogram
Abstract
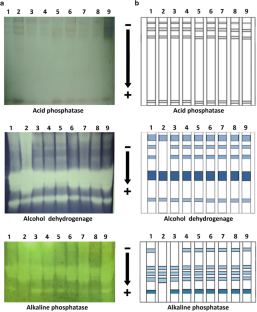
The present research work has been carried out for investigation of genetic diversity and phenetic relationship among nine Bangladeshi cultivars of cucumber (Cucumis sativus L.) using some isozymes and protein profiling by SDS-PAGE. Seventeen isozymes including acid phosphatase (ACP), alcohol dehydrogenase (ADH), alkaline phosphatase (ALP), amino peptidase (AMP), aspartate amino transferase (AAT), β-galactosidase (GAL), β-glucosidase (GLU), catalase (CAT), esterases (EST), formate dehydrogenase (FDH), fructose bisphosphatase (FBP), fumarate hydratase (FUM), hexokinase (HEX), malate dehydrogenase (MDH-NAD+), peroxidase (PRX), superoxide dismutase (SOD) and tyrosinase (TYR) were assayed. A total of 48 bands were detected resolved in 17 isozymes, out of which 21 were polymorphic. No polymorphism has been found in acid phosphatase, β-galactosidase, formate dehydrogenase, fumarate hydratase and peroxidase. A total of 23 protein bands were resolved, out of which 22 were polymorphic. Seven isozymes banding patterns were observed in Superoxide dismutase which was the highest among the 17 enzyme systems. Dendrograms and three-dimensional plots based on isozymes, SDS-PAGE and combined dataset clearly separated almost all the cultivars. From the phonetic analysis, it was revealed that the cultivars ‘Alavy-1’, ‘Baromasse’, ‘Raj-1’ and ‘Dinajpuri’ are somewhat more closely related than the other cultivars. The cultivar ‘Green king’ produced six cultivar specific bands in SDS-PAGE. This study will help successful breeding programmes in cucumber in Bangladesh.
References
- Ahmad M, McNeil DL (1996) Comparison of crossability, RAPD, SDS-PAGE and morphological markers for revealing genetic relationships within and among Lens species. Theor Appl Genet 93:788–793
- Ahmed MF, Iqbal M, Masood MS, Rabbani MA, Munir M (2010) Assessment of genetic diversity among Pakistani wheat (Triticum aestivum L.) advanced breeding lines using RAPD and SDS-PAGE. Electron J Biotechnol 13:1
- Amin ABMR, Rashid MM, Meah MB (2009) Efficacy of garlic tablet to control seed-borne fungal pathogens of cucumber. J Agric Rural Dev 7:135–138
- Anwar R, Masood S, Khan MA, Nasim S (2003) The high-molecular-weight glutenin subunit composition of wheat (Triticum aestivum L.) landraces from Pakistan. Pak J Bot 35:61–68
- Barman A, Singh DK, Maurya SK (2013) SDS-PAGE based protein profiling and diversity assessment of indigenous germplasms of phoot (Cucumis melo L. var momordica Duth & Full.). Prog Hort 45:169–173
- Bhandari AP, Sukanya DH, Ramesh CR (2006) Application of isozyme data in fingerprinting Napier grass (Pennisetum purpureum Schum.) for germplasm management. Genet Resour Crop Evol 53:253–264
- Bhat TM, Kudesia R (2011) Evaluation of genetic diversity in five different species of family Solanaceae using cytological characters and protein profiling. Genet Eng Biotechnol J 2011: GEBJ-20
- Bringhurst RS, Arulsekar S, Hancock JF Jr, Voth V (1981) Electrophoretic characterization of strawberry cultivars. J Am Soc Hortic Sci 106:684–687
- Chen J-F, Staub JE, Jiang J (1998) A reevaluation of karyotype in cucumber (Cucumis sativus L.). Genet Resour Crop Evol 45:301–305
- Chu Y-F, Sun J, Wu X, Liu RH (2002) Antioxidant and antiproliferative activities of common vegetables. J Agric Food Chem 50:6910–6916
- Colby LW, Peirce LC (1988) Using an isozyme marker to identify doubled haploids from anther culture of asparagus. HortScience 23:761–763
- Crawford DJ (1983) Phylogenetic and systematic inferences from electrophoretic studies. In: Tanksley SD, Orton TJ (eds) Developments in plant genetics and breeding, vol 1. Part A: isozymes in plant genetics and breeding, Part A. Elsevier, Amsterdam, pp 257–287
- Dar AA, Mahajan R, Lay P, Sharma S (2017) Genetic diversity and population structure of Cucumis sativus L. by using SSR markers. 3 Biotech 7:307
- del Río LA, Corpas FJ, López-Huertas E, Palma JM (2018) Plant superoxide dismutases: function under abiotic stress conditions. In: Gupta D, Palma J, Corpas F (eds) Antioxidants and antioxidant enzymes in higher plants. Springer, Cham, pp 1–26
- DeWald MG, Moore GA, Sherman WB (1988) Identification of pineapple cultivars by isozyme genotypes. J Am Soc Hortic Sci 113:935–938
- Eeswara JP, Peiris BCN (2001) Isoenzymes as markers for identification of Mungbean (Vigna radiata (L) Wilcezk). Seed Sci Technol 29:249–254
- Galani S, Naz F, Soomro F, Jamil I, Zia-ul-hassan Azhar A, Ashraf A (2011) Seed storage protein polymorphism in ten elite rice (Oryza sativa L.) genotypes of Sindh. Afr J Biotechnol 10:1106–1111
- Geburek T, Scholz F, Knabe W, Vornweg A (1987) Genetic studies by isozyme gene loci on tolerance and sensitivity in an air polluted Pinus sylvestris field trial. Silv Genet 36:49–53
- Ghafoor A, Ahmad Z, Qureshi AS, Bashir M (2002) Genetic relationship in Vigna mungo (L.) Hepper and V. radiata (L.) R. Wilczek based on morphological traits and SDS-PAGE. Euphytica 123:367–378
- Gorman MB, Kiang YT (1977) Variety-specific electrophoretic variants of four soybean enzymes. Crop Sci 17:963–965
- Guo S, Zheng Y, Joung J-G, Liu S, Zhang Z, Crasta OR, Sobral BW, Xu Y, Huang S, Fei Z (2010) Transcriptome sequencing and comparative analysis of cucumber flowers with different sex types. BMC Genomics 11:384
- Han Y, Zhang Z, Liu C, Liu J, Huang S, Jiang J, Jin W (2009) Centromere repositioning in cucurbit species: implication of the genomic impact from centromere activation and inactivation. Proc Natl Acad Sci USA 106:14937–14941
- Hasan R, Hossain MK, Alam N, Bashar A, Islam S, Tarafder MJA (2015) Genetic divergence in commercial cucumber (Cucumis Sativus L.) genotypes. Bangladesh J Bot 44:201–207
- Hauser LA, Crovello TJ (1982) Numerical analysis of generic relationships in Thelypodieae (Brassicaceae). Syst Bot 7:249–268
- Hirai M, Kozaki I, Kajiura I (1986) Isozyme analysis and phylogenic relationship of citrus. Jpn J Breed 36:377–389
- Horejsi T, Staub JE (1999) Genetic variation in cucumber (Cucumis sativus L.) as assessed by random amplified polymorphic DNA1. Genet Resour Crop Evol 46:337–350
- Huang S, Li R, Zhang Z, Li L, Gu X, Fan W, Lucas WJ, Wang X, Xie B, Ni P et al (2009) The genome of the cucumber, Cucumis sativus L. Nat Genet 41:1275–1281
- Hunter RL, Markert C (1957) Histochemical demonstration of enzymes separated by zone electrophoresis in starch gels. Science 125:1294–1295
- Iqbal SM, Ghafoor A, Ayub N (2005) Relationship between SDS-PAGE markers and Ascochyta blight in chickpea. Pak J Bot 37:87–96
- Isshiki T, Mochizuki N, Maeda T, Yamamoto M (1992) Characterization of a fission yeast gene, gpa2, that encodes a Gα subunit involved in the monitoring of nutrition. Genes Dev 6:2455–2462
- Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat 44:223–270
- Jat GS, Munshi AD, Behera TK, Tomar BS (2016) Combining ability estimation of gynoecious and monoecious hybrids for yield and earliness in cucumber (Cucumis sativus). Ind J Agric Sci 86:399–403
- Javaid A, Ghafoor A, Anwar R (2004) Seed storage protein electrophoresis in groundnut for evaluating genetic diversity. Pak J Bot 36:25–29
- Kar D, Pattanaik PK, Acharya L, Panda MK, Sathapathy K, Kuanar A, Mishra B (2014) Assessment of genetic diversity among some elite cultivars of ginger (Zingiber officinale Rosc.) using isozyme and protein markers. Braz J Bot 37:469–479
- Knerr LD, Staub JE, Holder DJ, May BP (1989) Genetic diversity in Cucumis sativus L. assessed by variation at 18 allozyme coding loci. Theor Appl Genet 78:119–128
- Kroon GH, Custers JBM, Kho YO, den Nijs APM, Varekamp HQ (1979) Interspecific hybridization in Cucumis (L.). I. Need for genetic variation, biosystematic relations and possibilities to overcome crossability barriers. Euphytica 28:723–728
- Kupper RS, Staub JE (1988) Combining ability between lines of Cucumis sativus L. and Cucumis sativus var. hardwickii (R.) Alef. Euphytica 38:197–210
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
- Lv J, Qi J, Shi Q, Shen D, Zhang S, Shao G, Li H, Sun Z, Weng Y, Shang Y et al (2012) Genetic diversity and population structure of cucumber (Cucumis sativus L.). PLoS ONE 7:e46919
- Madina MH, Rahman MS, Deb AC, Choi YH, Kim MR, Shin J, Yoo JC (2015) Polymorphism Assessment of Six Lentil (Lens culinaris Medik.) Genotypes Using Isozyme. J Chosun Natl Sci 8:117–127
- Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
- Markert CL, Møller F (1959) Multiple forms of enzymes: tissue, ontogenetic, and species specific patterns. Proc Natl Acad Sci USA 45:753–763
- Meglic V, Staub JE (1996) Inheritance and linkage relationships of isozyme and morphological loci in cucumber (Cucumis sativus L.). Theor Appl Genet 92:865–872
- Meglic V, Serquen F, Staub JE (1996) Genetic diversity in cucumber (Cucumis sativus L.): I. A reevaluation of the US germplasm collection. Genet Resour Crop Evol 43:533–546
- Milner JA (2000) Functional foods: the US perspective. Am J Clin Nutr 71:1654S–1659S
- Mirali N, El-Khouri S, Rizq F (2007) Genetic diversity and relationships in some Vicia species as determined by SDS-PAGE of seed proteins. Biol Plant 51:660–666
- Mukherjee A, Sikdar B, Ghosh B, Banerjee A, Ghosh E, Bhattacharya M, Roy SC (2013) Isozyme variation in some economically important species of the Genus Allium L. (Alliaceae). J Herbs Spices Med Plants 19:297–312
- Mündges H, Köhler W, Friedt W (1990) Identification of rape seed cultivars (Brassica napus) by starch gel electrophoresis of enzymes. Euphytica 45:179–187
- Murphy RW, Sites JW, Buth DG, Haufler CH (1990) Proteins: isozyme electrophoresis. In: Hillis DH, Moritz C (eds) Molecular systematics. Sinauer Associates Inc, Publishers, Sunderland, pp 45–126
- Nethra N, Prasad SR, Vishwanath K, Dhanraj KN, Gowda R (2007) Identification of rice hybrids and their parental lines based on seed, seedling characters, chemical tests and gel electrophoresis of total soluble proteins. Seed Sci Technol 35:176–186
- Perl-Treves R, Zamir D, Navot N, Galun E (1985) Phylogeny of Cucumis based on isozyme variability and its comparison with plastome phylogeny. Theor Appl Genet 71:430–436
- Peterson CE (1975) Plant introductions in the improvement of vegetable cultivars. HortScience 10:575–579
- Rahman MM, Nito N (1994) Phylogenetic relationships among the “true Citrus fruit trees” by glutamate oxaloacetate transaminase isozymes analysis. J Jpn Soc Hortic Sci 62:755–760
- Ren Y, Zhang Z, Liu J, Staub JE, Han Y, Cheng Z, Li X, Lu J, Miao H, Kang H et al (2009) An integrated genetic and cytogenetic map of the cucumber genome. PLoS ONE 4:e5795
- Rohlf FJ (2009) NTSYSpc: numerical taxonomy system. ver. 2.21c. Exeter Software, Setauket
- Sadia M, Malik SA, Rabbani MA, Pearce SR (2009) Electrophoretic characterization and the relationship between some Brassica species. Electronic J Biol 5:1–4
- Samec P, Pošvec Z, Stejskal J, Našinec V, Griga M (1998) Cultivar identification and relationships in Pisum sativum L. based on RAPD and isoenzymes. Biol Plant 41:39–48
- Sánchez-Escribano E, Ortiz JM, Cenis JL (1998) Identification of table grape cultivars (Vitis vinifera L.) by the isoenzymes from the woody stems. Genet Resour Crop Evol 45:173–179
- Schulze J, Balko C, Zellner B, Koprek T, Hänsch R, Nerlich A, Mendel RR (1995) Biolistic transformation of cucumber using embryogenic suspension cultures: long-term expression of reporter genes. Plant Sci 112:197–206
- Shuaib M, Jamal M, Akbar H, Khan I, Khalid R (2010) Evaluation of wheat by polyacrylamide gel electrophoresis. Afr J Biotechnol 9:243–247
- Sikdar B, Bhattacharya M, Mukherjee A, Banerjee A, Ghosh E, Ghosh B, Roy SC (2010) Genetic diversity in important members of Cucurbitaceae using isozyme, RAPD and ISSR markers. Biol Plant 54:135–140
- Singh NP, Matta NK (2008) Variation studies on seed storage proteins and phylogenetics of the genus Cucumis. Plant Syst Evol 275:209–218
- Singh DK, Tewari R, Singh NK, Singh SS (2016) Genetic diversity cucumber using inter simple sequence repeats (ISSR). Transcriptomics 4:129
- Soost RK, Williams TE, Torres AM (1980) Identification of nucellar and zygotic seedlings of Citrus with leaf isozymes. HortScience 15:728–729
- Srivastava D, Khan NA, Shamim M, Yadav P, Pandey P, Singh KN (2014) Assessment of the genetic diversity in bottle gourd (Lagenaria siceraria [Molina] Standl.) genotypes using SDS-PAGE and RAPD Markers. Natl Acad Sci Lett 37:155–161
- Staub JE, Fredrick L (1988) Inheritance of three enzyme loci in cucumber. HortScience 23:757–758
- Staub JE, Serquen FC, McCreight JD (1997) Genetic diversity in cucumber (Cucumis sativus L.): III. An evaluation of Indian germplasm. Genet Resour Crop Evol 44:315–326
- Staub JE, Serquen FC, Horejsi T, Chen J (1999) Genetic diversity in cucumber (Cucumis sativus L.): IV. An evaluation of Chinese germplasm. Genet Resour Crop Evol 46:297–310
- Sultana T, Ghafoor A (2008) Genetic diversity in ex situ conserved Lens culinaris for botanical descriptors, biochemical and molecular markers and identification of landraces from indigenous genetic resources of Pakistan. J Integr Plant Biol 50:484–490
- Tanksley SD, Jones RA (1981) Effects of O2 stress on tomato alcohol dehydrogenase activity: description of a second ADH coding gene. Biochem Genet 19:397–409
- Wang Y-H, Joobeur T, Dean RA, Staub JE (2007) Cucurbits. In: Kole C (ed) Genome mapping and molecular breeding in plants, Vegetables, vol 5. Springer, Berlin, pp 315–329
- Weeden NF, Lamb RC (1987) Genetics and linkage analysis of 19 isozyme loci in apple. J Am Soc Hortic Sci 110:509–515
- Weeden NF, Provvidenti R, Marx GA (1984) An isozyme marker for resistance to bean yellow mosaic virus in Pisum sativum. J Hered 75:411–412
- Wendel JF, Parks CR (1982) Genetic control of isozyme variation in Camellia japonica L. J Hered 73:197–204
- Wendel JF, Weeden NF (1989) Visualization and interpretation of plant isozymes. In: Soltis DE, Soltis PS (eds) Isozymes in plant biology. Chapman and Hall, London, pp 5–45
- Woods S, Thurman DA (1976) The use of seed acid phosphatases in the determination of the purity of F1 hybrid Brussels sprout seed. Euphytica 25:707–712
- Yadav H, Maurya SK, Bhat L (2015) SDS-Page based seed protein profiling and diversity assessment of indigenous genotypes of ridge gourd (Luffa acutangula (L.) Roxb.). Int J Agric Environ Biotechnol 8:501–510
Author Information
Professor Joarder DNA & Chromosome Research Lab., Department of Genetic Engineering and Biotechnology, University of Rajshahi, Rajshahi, Bangladesh