Development of advance pearl millet lines tolerant to terminal drought stress using marker-assisted selection
Rani Asha, Taunk Jyoti, Jangra Sumit, Yadav Ram C., Yadav Neelam R., Yadav Devvart, Yadav H. P.
Research Articles | Published: 14 August, 2021
First Page: 63
Last Page: 73
Views: 4397
Keywords: Pearl millet, Drought tolerance, Marker-assisted backcrossing, QTLs, SSRs
Abstract
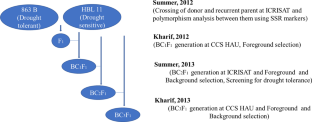
HHB 226 is a popular, high-yielding pearl millet hybrid cultivated widely in the Northern part of India. Drought stress at post-flowering stage is one of the major constraints in realizing potential yields of this hybrid. Developing drought-tolerant pearl millet hybrid HHB 226 is priority for pearl millet breeders. Transferring the drought-tolerant quantitative trait loci (QTLs) from donor source (863 B) to male parent of HHB 226 (HBL 11) was undertaken to improve HHB 226. The drought-tolerant donor source, 863 B was backcrossed with recurrent parent (HBL 11) up to BC3F1 generation and drought-tolerant QTLs were transferred through marker-assisted backcrossing (MABC) strategy. Four polymorphic simple sequence repeat (SSR) markers linked to drought-tolerant QTLs were used to select the plants with drought-tolerant QTLs in each generation. In BC3F1 generation, four such plants were selected. Background selection was done in BC2F1 and BC3F1 generations for recovery of recurrent parent genome. Thirty-two background markers, found polymorphic out of 64 primers screened, were further used for background selection. About 80% of recurrent parent genome has been recovered in selected plants of BC3F1 generation. The results demonstrated that drought-tolerant QTLs transferred through MAS successfully improved drought tolerance in HHB 226.
References
Allouis S, Qi X, Lindup S, Gale MD, Devos KM (2001) Constructions of BAC library of pearl millet, Pennisetum glaucum. Theor Appl Genet 102:1200–1205
Ambawat S, Singh S, Shobhit, Meena RC, Satyavathi TC (2020) Biotechnological applications for improvement of the pearl millet crop. In: Punia S, Siroha AK, Sandhu KS, Gahlawat SK, Kaur M (eds) Pearl millet properties, functionality and its applications. CRC Press, New York. https://doi.org/10.1201/978042933173 (2)
Babu R, Nair SK, Prasanna BA, Gupta HS (2004) Integrated marker-assisted selection in crop breeding: prospects and challenges. Curr Sci 87(5):607–616
Benchimol L, de Souza C, Anete Pereira S (2005) Microsatellite-assisted backcross selection in maize. Genet Mol Biol 28(4):789–797
Bidinger FR, Mahalakshmi V, Rao GDP (1987) Assessment of drought resistance in pearl millet [Pennisetum americanum (L.) Leeke]. I. Factors affecting yield under stress. Aust J Agric Res 38:37–48
Bidinger FR, Serraj R, Rizvi SMH, Howarth CJ, Yadav RS, Hash CT (2005) Field evaluation of drought tolerance QTL effects on phenotype and adaptation in pearl millet [Pennisetum glaucum (L.) R. Br.] topcross hybrids. Field Crop Res 94:14–32
Budak H, Pedraza F, Cregan PB, Baenziger P, Dweikat S (2003) Development and utilization of SSRs to estimate the degree of genetic relationships in a collection of pearl millet germplasm. Crop Sci 43:2284–2290
Chowdhury J, Karim M, Khaliq Q, Solaiman A, Ahmed J (2016) Genotypic variations in growth, yield and yield components of soybean genotypes under drought stress conditions. Bangladesh J Agr Res 40(12):537–550
Darkwa K, Ambachew D, Mohammed H, Asfaw A, Blair MW (2016) Evaluation of common bean (Phaseolus vulgaris L.) genotypes for drought stress adaptation in Ethiopia. Crop J 4(5):367–376
Debieu M, Sine B, Passot S, Grondin A, Akata E, Gangashetty P, Vadez V, Gantet P, Fonce D, Cournac L, Hash CT, Kane NA, Vigouroux Y, Laplaze L (2018) Response to early drought stress and identification of QTLs controlling biomass production under drought in pearl millet. PLoS One 13(10):e0201635. https://doi.org/10.1371/journal.pone.0201635
Dudhate A, Shinde H, Tsugama D, Liu S, Takano T (2018) Transcriptomic analysis reveals the differentially expressed genes and pathways involved in drought tolerance in pearl millet [Pennisetum glaucum (L.) R. Br]. PLoS One 13(4):e0195908. https://doi.org/10.1371/journal.pone.0195908
Gandhi D (2007) UAS scientist develops first drought tolerant rice. The Hindu. http://www.thehindu.com/2007/11/17/stories/2007111752560500.htm. Accessed 30 Mar 2021
Hash CT, Sharma A, Kolesnikova-Allen MA, Singh SD, Thakur RP, Bhasker Raj AG, Ratnaji Rao MNV, Nijhawan DC, Beniwal CR, Sagar P, Yadav HP, Yadav YP, Srikant Bhatnagar SK, Khairwal IS, Howarth CJ, Cavan GP, Gale MD, Liu C, Devos KM, Breese WA, Witcombe JR (2005) Teamwork delivers biotechnology products to Indian small-holder crop-livestock producers: Pearl millet hybrid “HHB 67 Improved” enters seed delivery pipeline. J SAT Agr Res 2(1):1–3
Hash CT, Yadav RS, Cavan GP, Howarth CJ, Liu H, Qi X, Sharma A, Kolesnikova-Allen MA, Bidinger FR, Witcombe JR (2000) Marker-assisted backcrossing to improve drought tolerance in pearl millet. In: Ribaut JM, Poland D (Eds.) Molecular Approaches for the Genetic Improvement of Cereals for Stable Production in Water-Limited Environments. A Strategic Planning Workshop held at International Maize and Wheat Improvement Center (CIMMYT), Mexico, pp 114–119
Hospital F (2005) Selection in backcross programmes. Phil Trans R Soc B 360:1503–1511
Howarth CJ, Yadav RS (2002) Successful MAS for drought tolerance & disease resistance in pearl millet. IGER Innovation, pp 18–21
Iftekharuddaula KM, Salam MA, Newaz MA, Ahmed HU, Collard BC, Septiningsih EM, Sanchez DL, Pamplona AM, Mackill DJ (2012) Comparison of phenotypic versus marker-assisted background selection for the SUB1 QTL during backcrossing in rice. Breeding Sci 62:216–222
Jaiswal S, Antala TJ, Mandavia MK, Chopra M, Jasrotia RS, Tomar RS, Kheni J, Angadi UB, Iquebal MA, Golakia BA, Rai A, Kumar D (2018) Transcriptomic signature of drought response in pearl millet (Pennisetum glaucum (L.) and development of web-genomic resources. Sci Rep 8:3382. https://doi.org/10.1038/s41598-018-21560-1
Kamaluddin K, Kiran MA, Ali U, Abdin A, Zargar MZ, Ahmad MY, Sofi S, Gulzar S (2017) Molecular markers and marker-assisted selection in crop plants. In: Abdin M, Kiran U, Kamaluddin, Ali A (eds) Plant biotechnology: principles and application. Springer, Singapore, pp 295–328
Khairwal IS, Hash CT (2007) “HHB 67-Improved”—The first product of marker-assisted crop breeding in India. Asia-Pacific Consortium on Agricultural Biotechnology (APCoAB) e-News. http://www.apcoab.org/special_news.html. Accessed 30 Mar 2021
Kumar A, Elston J (1992) Genotypic differences in leaf water relations between B. juncea and B. napus. Ann Bot 70:3–9
Kumar S, Hash CT, Singh G, Nepolean T, Srivastava RK (2021) Mapping QTLs for important agronomic traits in an Iniadi-derived immortal population of pearl millet. Biotechnol Notes 2:26–32
Kuroda Y, Kaga A, Tomooka N, Yano H, Takada Y, Kato S, Vaughan D (2013) QTL affecting fitness of hybrids between wild and cultivated soybeans in experimental fields. Ecol Evol 3(7):2150–2168
Liu CJ, Witcombe JR, Pittaway TS, Nash M, Hash CT, Busso CS, Gale MD (1994) An RFLP-based genetic map of pearl millet (Pennisetum glaucum). Theor Appl Genet 89:481–487
Malik S (2015) Pearl millet-nutritional value and medicinal uses! IJARIIE 1:414–418
Mariac C, Luong V, Kapran I, Mamadou A, Sagnard F, Deu M, Chantereau J, Gerard B, Ndjeunga J, Bezançon G, Pham JL, Vigouroux Y (2006) Diversity of wild and cultivated pearl millet accessions (Pennisetum glaucum [L.] R. Br.) in Niger assessed by microsatellite markers. Theor Appl Genet 114:49–58
Meher SP, Ashok Reddy K, Manohar Rao D (2018) Effect of PEG-6000 imposed drought stress on RNA content, relative water content (RWC), and chlorophyll content in peanut leaves and roots. Saudi J Biol Sci 25(2):285–289
Muthamilarasan M, Dhaka A, Yadav R, Prasad M (2016) Plant science exploration of millet models for developing nutrient rich graminaceous crops. Plant Sci 242:89–97
Pandey MK, Rani NS, Sundaram RM, Laha GS, Madhav MS, Rao KS, Sudharshan S, Hari Y, Varaprasad GS, Subba Rao LV, Suneetha K, Sivaranjani AK, Viraktamath BC (2013) Improvement of two traditional Basmati rice varieties for bacterial blight resistance and plant stature through morphological and marker-assisted selection. Mol Breed 31:239–246
Qi X, Pittaway TS, Lindup S, Liu H, Waterman E, Padi FK, Hash CT, Zhu J, Gale MD, Devos KM (2004) An integrated genetic map and a new set of simple sequence repeat marker for pearl millet, Pennisetum glaucum. Theor Appl Genet 109:1485–1493
Rajpurohit D, Kumar R, Kumar M, Paul P, Awasthi AA, Basha PO, Puri A, Jhang T, Singh K, Dhaliwal HS (2010) Pyramiding of two bacterial blight resistance and a semi dwarfing gene in Type 3 Basmati using marker-assisted selection. Euphytica 178:111–126
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphism in barley: Mendelian inheritance, chromosomal-location and population dynamics. Proc Natl Acad Sci USA 81:8014–8018
Sehgal D, Rajaram V, Armstead IP, Vadez V, Yadav YP, Hash CT, YadaV RS (2012) Integration of gene-based markers in a pearl millet genetic map for identification of candidate genes underlying drought tolerance quantitative trait loci. BMC Plant Biol 12:9
Sehgal D, Skot L, Singh R, Srivastava RK, Das SP, Taunk J, Sharma PC, Pal R, Raj B, Hash CT, Yadav RS (2015) Exploring potential of pearl millet germplasm association panel for association mapping of drought tolerance traits. PLoS One 10(5):e0122165. https://doi.org/10.1371/journal.pone.0122165
Senthivel S, Jayashree B, Mahalakshmi V, Sathish KP, Nakka S, Nepolean T, Hash CT (2008) Developing and mapping of SSR markers for pearl millet from data mining of expressed sequence tags. BMC Plant Biol 8:119–127
Serba DD, Yadav RS (2016) Genomic tools in pearl millet breeding for drought tolerance: status and prospects. Front Plant Sci 7:1724. https://doi.org/10.3389/fpls.2016.01724
Serraj R, Hash CT, Rizvi SMH, Sharma A, Yadav RS, Bidinger FR (2005) Recent advances in marker-assisted selection for drought tolerance in pearl millet. Plant Prod Sci 8:334–337
Shivhare R, Lata C (2017) Exploration of genetic and genomic resources for abiotic and biotic stress tolerance in pearl millet. Front Plant Sci 7:2069
Shivhare R, Asif MH, Lata C (2020a) Comparative transcriptome analysis reveals the genes and pathways involved in terminal drought tolerance in pearl millet. Plant Mol Biol 103:639–652
Shivhare R, Lakhwani D, Asif MH, Chauhan PS, Lata C (2020b) De novo assembly and comparative transcriptome analysis of contrasting pearl millet (Pennisetum glaucum L.) genotypes under terminal drought stress using illumina sequencing. Nucleus 63:341–352
Soni P (2020) Understanding the mechanism of drought tolerance in pearl millet. In: Kuila A (ed) Plant stress biology: progress and prospects of genetic engineering. Apple Academic Press, New York. https://doi.org/10.1201/9781003055358
Sundarum RM, Vishnupriya MR, Biradar SK, Laha GS, Reddy GA, Rani NS, Sharma NP, Sonti RV (2008) Marker-assisted introgression of bacterial blight resistance in Samba Mashuri, an elite indica rice variety. Euphytica 160(3):411–422
Supriya A, Senthievel S, Nepolean T, Eshwar K, Rajaram V, Shaw R, Hash CT, Kilian A, Yadav RC, Narasu ML (2011) Development of a molecular linkage map of pearl millet integrating DArT & SSR markers. Theor Appl Genet. https://doi.org/10.1007/s00122-011-1580-1
Tako E, Reed SM, Budiman J, Hart JJ, Glahn RP (2015) Higher iron pearl millet (Pennisetum glaucum L.) provides more absorbable iron that is limited by increased polyphenolic content. Nutr J 14(1):1–11
Tanksley SD, Nelson JC (1996) Advanced backcross QTLs analysis: a method for the simultaneous discovery and transfer of valuable QTLs from unadapted germplasm into elite breeding lines. Theor Appl Genet 92:191–203
Taunk J, Rani A, Yadav NR, Yadav D, Yadav RC, Raj K, Kumar R, Yadav HP (2018) Molecular breeding of ameliorating commercial pearl millet hybrid for downy mildew resistance. J Genet 97(5):1241–1251
Van Berloo R (1999) GGT: software for display of graphical genotypes. J Hered 90:328–329
Varshney RK, Bansal KC, Aggarwal PK, Datta SK, Crauford PQ (2011) Agricultural biotechnology for crop improvement in a variable climate: hope or hype? Trends Plant Sci 16(7):363–371
Varshney RK, Shi C, Thudi M, Mariac C, Wallace J, Qi P, Zhang H, Zhao Y, Wang X, Rathore A, Srivastava RK, Chitikineni A, Fan G, Bajaj P, Punnuri S, Gupta SK, Wang H, Jiang Y, Couderc M, Katta MAVSK, Paudel DR, Mungra KD, Chen W, Harris-Shultz KR, Garg V, Desai N, Doddamani D, Kane NA, Conner JA, Ghatak A, Chaturvedi P, Subramaniam S, Yadav OP, Berthouly-Salazar C, Hamidou F, Wang J, Liang X, Clotault J, Upadhyaya HD, Cubry P, Rhoné B, Gueye MC, Sunkar R, Dupuy C, Sparvoli F, Cheng S, Mahala RS, Singh B, Yadav RS, Lyons E, Datta SK, Hash CT, Devos KM, Buckler E, Bennetzen JL, Paterson AH, Ozias-Akins P, Grando S, Wang J, Mohapatra T, Weckwerth W, Reif JC, Liu X, Vigouroux Y, Xu X (2017) Pearl millet genome sequence provides a resource to improve agronomic traits in arid environments. Nat Biotechnol 35(10):969–976
Vetriventhan M, Nirmalakumari A, Ganapathy S (2008) Heterosis for grain yield components in pearl millet (Pennisetum glaucum (L.) R. Br.). WJAS 4(5):657–660
Xu Y, Crouch HJ (2008) Marker-assisted selection in plant breeding: from publications to practice. Crop Sci 48:391–407
Yadav RS, Hash CT, Bidinger FR, Cavan GP, Howarth CJ (2002) QTL associated with traits determining grain and stover yield in pearl millet under terminal drought stress condition. Theor Appl Genet 104:67–83
Yadav RS, Hash CT, Bidinger FR, Devos KM, Howarth CJ (2004) Genomic regions associated with grain yield and aspects of post-flowering drought tolerance in pearl millet across stress environments and testers background. Euphytica 136:265–277
Yadav RS, Sehgal D, Vadez V (2011) Using genetic mapping and genomics approaches in understanding and improving drought tolerance in pearl millet. J Exp Bot 62(2):397–408
Author Information
Department of Molecular Biology, Biotechnology, and Bioinformatics, CCS Haryana Agricultural University, Hisar, India