Development of tetra-primer ARMS-PCR to genotype the G1178A SNP on the Cucumis sativus Bitterness gene
Short Communications | Published: 16 November, 2024
First Page: 2099
Last Page: 2104
Views: 2107
Keywords: Bitterness, Codominant marker, Cucumber, Heterozygous genotype, Single-nucleotide polymorphism (SNP), Tetra-primer ARMS-PCR
Abstract
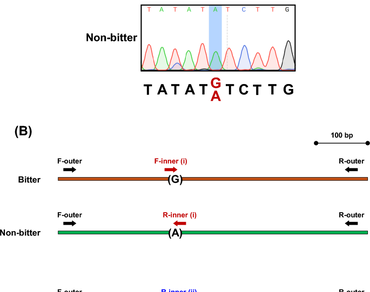
Cucumber can be considered one of the most popular fruity vegetables worldwide. However, numerous cultivated cucumber varieties exhibit to produce cucurbitacins that lead to bitter taste in this crop species. Previous efforts uncovered one single-nucleotide polymorphism (SNP) that obviously links to the bitterness trait in cucumbers. The SNP was identified as a nucleotide substitution at position 1178 within the coding sequence of the cucumber Bitterness gene (G1178A). Earlier, a few real-time PCR-based techniques were developed to genotype this type of SNP in cucumbers. In this work, we aim to establish a tetra-primer ARMS-PCR-based method to genotype the G1178A SNP in cucumbers. The present method shows to efficiently identify the SNP in both homozygous and heterozygous genotypes. It can be potentially applied in further cucumber breeding projects aiming to generate new free-bitterness cucumber varieties.
References
Andeweg JM, De Bruyn JW (1959) Breeding of non-bitter cucumbers. Euphytica 8(1):13–20. https://doi.org/10.1007/BF00022084
Chen J, Xu X, Dalhaimer P, Zhao L (2022) Tetra-primer amplification-refractory mutation system (ARMS)—PCR for genotyping mouse Leptin gene mutation. Animals 12(19):2680. https://doi.org/10.3390/ani12192680
Draelos ZD (2014) Facial skin care products and cosmetics. Clin Dermatol 32(6):809–812. https://doi.org/10.1016/j.clindermatol.2014.02.020
Horie H, Ito H, Ippoushi K, Azuma K, Sakata Y, Igarashi I (2007) Cucurbitacin C—bitter principle in cucumber plants. Japan Agricult Res Quar: JARQ 41(1):65–68. https://doi.org/10.6090/jarq.41.65
Huang S, Li R, Zhang Z, Li L, Gu X, Fan W, Lucas WJ, Wang X, Xie B, Ni P, Ren Y, Zhu H, Li J, Lin K, Jin W, Fei Z, Li G, Staub J, Kilian A, Li S (2009) The genome of the cucumber, Cucumis sativus L. Nat Genetics 41(12):1275–1281. https://doi.org/10.1038/ng.475
Ke-xin Y, Xiang C, Qing-qing H, Yi-an Y, Xiao-ming W, Ai-chun X, Jian G, Feng G (2023) Development of a tetra-primer ARMS–PCR for identification of sika and red deer and their hybrids. Anal Sci 39(12):1947–1956. https://doi.org/10.1007/s44211-023-00405-6
Khan A, Mishra A, Hasan SM, Usmani A, Ubaid M, Khan N, Saidurrahman M (2022) Biological and medicinal application of Cucumis sativus Linn.—Review of current status with future possibilities. J Complement Integr Med 19(4):843–854. https://doi.org/10.1515/jcim-2020-0240
Kou M, Xu J, Li Q, Liu Y, Wang X, Tang W, Yan H, Zhang Y, Ma D (2017) Development of SNP markers using RNA-seq technology and tetra-primer ARMS-PCR in sweetpotato. J Integr Agric 16(2):464–470. https://doi.org/10.1016/S2095-3119(16)61405-3
Kundu A, Ghosh P, Bishayi B (2024) Vitexin along with verapamil downregulates efflux pump P-glycoprotein in macrophages and potentiate M1 to M2 switching via TLR4-NF-κB-TNFR2 pathway in lipopolysaccharide treated mice. Immunobiology 229(1):152767. https://doi.org/10.1016/j.imbio.2023.152767
Lawrie RD, Massey SE (2023) Agrigenomic Diversity Unleashed: current single nucleotide polymorphism genotyping methods for the agricultural sciences. Appl Biosci 2(4):565–585. https://doi.org/10.3390/applbiosci2040036
Li Y, Li Y, Yao Y, Li H, Gao C, Sun C, Zhuang J (2023) Potential of cucurbitacin as an anticancer drug. Biomed Pharmac 168:115707. https://doi.org/10.1016/j.biopha.2023.115707
Ma L, Wang Q, Zheng Y, Guo J, Yuan S, Fu A, Bai C, Zhao X, Zheng S, Wen C, Guo S, Gao L, Grierson D, Zuo J, Xu Y (2022) Cucurbitaceae genome evolution, gene function, and molecular breeding. Horticult Res. https://doi.org/10.1093/hr/uhab057
Matsui K, Mizuno N, Ueno M, Takeshima R, Yasui Y (2020) Development of co-dominant markers linked to a hemizygous region that is related to the self-compatibility locus (S) in buckwheat (Fagopyrum esculentum). Breed Sci 70(1):112–117. https://doi.org/10.1270/jsbbs.19129
Medrano RFV, de Oliveira CA (2014) Guidelines for the tetra-primer ARMS–PCR technique development. Mol Biotechnol. https://doi.org/10.1007/s12033-014-9734-4
Mukherjee PK, Nema NK, Maity N, Sarkar BK (2013) Phytochemical and therapeutic potential of cucumber. Fitoterapia 84:227–236. https://doi.org/10.1016/j.fitote.2012.10.003
Phillips DR, Rasbery JM, Bartel B, Matsuda SP (2006) Biosynthetic diversity in plant triterpene cyclization. Curr Opin Plant Biol 9(3):305–314. https://doi.org/10.1016/j.pbi.2006.03.004
Qing Z, Shi Y, Han L, Li P, Zha Z, Liu C, Liu X, Huang P, Liu Y, Tang Q, Zeng K, Zeng J, Zhou Y (2022) Identification of seven undescribed cucurbitacins in Cucumis sativus (cucumber) and their cytotoxic activity. Phytochemistry 197:113123. https://doi.org/10.1016/j.phytochem.2022.113123
Shang Y, Ma Y, Zhou Y, Zhang H, Duan L, Chen H, Zeng J, Zhou Q, Wang S, Gu W, Liu M, Ren J, Gu X, Zhang S, Wang Y, Yasukawa K, Bouwmeester HJ, Qi X, Zhang Z, Huang S (2014) Biosynthesis, regulation, and domestication of bitterness in cucumber. Science 346(6213):1084–1088. https://doi.org/10.1126/science.1259215
Swarup S, Cargill EJ, Crosby K, Flagel L, Kniskern J, Glenn KC (2021) Genetic diversity is indispensable for plant breeding to improve crops. Crop Sci 61(2):839–852. https://doi.org/10.1002/csc2.20377
Varela C, Melim C, Neves BG, Sharifi-Rad J, Calina D, Mamurova A, Cabral C (2022) Cucurbitacins as potential anticancer agents: new insights on molecular mechanisms. J Transl Med 20(1):630. https://doi.org/10.1186/s12967-022-03828-3
Venkatesh J, Song K, Lee J-H, Kwon J-K, Kang B-C (2018) Development of Bi gene-based SNP markers for genotyping for bitter-free cucumber lines. Hortic Environ Biotechnol 59(2):231–238. https://doi.org/10.1007/s13580-018-0029-8
Yang Z, Lo YT, Quan Z, He J, Chen Y, Faller A, Chua T, Wu HY, Zhang Y, Zou Q, Li F, Chang P, Swanson G, Shaw PC, Lu Z (2023) Application of a modified tetra-primer ARMS–PCR assay for rapid Panax species identity authentication in ginseng products. Sci Rep 13(1):14396. https://doi.org/10.1038/s41598-023-39940-7
Ye S (2001) An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res 29(17):88e–888. https://doi.org/10.1093/nar/29.17.e88
Zeng Y, Wang J, Huang Q, Ren Y, Li T, Zhang X, Yao R, Sun J (2021) Cucurbitacin IIa: a review of phytochemistry and pharmacology. Phytother Res 35(8):4155–4170. https://doi.org/10.1002/ptr.7077
Zhang J, Yang J, Zhang L, Luo J, Zhao H, Zhang J, Wen C (2020) A new SNP genotyping technology target SNP-seq and its application in genetic analysis of cucumber varieties. Sci Rep 10(1):5623. https://doi.org/10.1038/s41598-020-62518-6
Zhou Y, Ma Y, Zeng J, Duan L, Xue X, Wang H, Lin T, Liu Z, Zeng K, Zhong Y, Zhang S, Hu Q, Liu M, Zhang H, Reed J, Moses T, Liu X, Huang P, Qing Z, Huang S (2016) Convergence and divergence of bitterness biosynthesis and regulation in Cucurbitaceae. Nat Plant 2(12):16183. https://doi.org/10.1038/nplants.2016.183
Author Information
Faculty of Biotechnology, Ho Chi Minh City Open University, Ho Chi Minh City, Vietnam