Genetic diversity in Groundnut (Arachis hypogaea. L) genotypes varying in maturity duration
Vishnuprabha R. Sangeetha, Manonmani S., Rajendran L., Selvakumar T.
Short Communications | Published: 11 November, 2022
First Page: 1550
Last Page: 1556
Views: 3604
Keywords: Legume crop, SSR markers, D2 analysis, NTSys, Primer, PIC value
Abstract
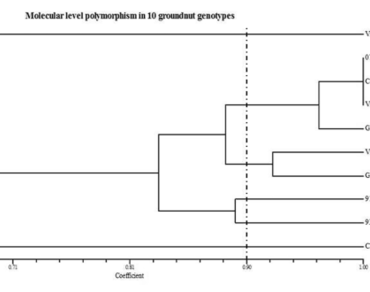
Groundnut (Arachis hypogaea L.) is a legume crop, a member of the Fabaceae family cultivated for food and one of the cash crop across the world. Selected 10 genotypes viz., CO 7, ICGV 07222, VRI 6, VRI8, GPBD 4, VRI 3, Chico, Gangapuri, ICGV 91,114 and ICGV 93,468 varying in maturity duration from short to long were clustered at molecular level through SSR markers. The selected genotypes were clustered into four clusters by D2 analysis and six clusters through NTSys analysis, grouping the early maturing varieties and high yielding varieties, separately. The trait kernel yield per plant contributed the maximum towards divergence among the selected genotypes of 38.50% followed by hundred pods weight with a contribution of 12.6%. The analysis of SSR markers on 10 genotypes, the primer GM 2637 exhibited maximum number of alleles (7) with higher PIC value (0.79).
References
Akhtar S, Shahzad A, Bangash N, Arshad M, Ahmed I (2014) Morpho-physiological and genetic diversity of groundnut (Arachis hypogaea L.) genotypes under iron deficiency stress. Pakistan Journal of Agricultural Sciences, 51(4): 953–961
Bhakal M, Lal G (2015) Studies on genetic diversity in groundnut (Arachis hypogaea L.) germplasm. Journal of Plant Science & Research, 2(2):1–4
Chaudhari S, Khare D, Sundravadana S, Manohar S, Variath MT (2017) Capturing genetic diversity in genomic selection panel of groundnut for foliar disease resistance, yield and nutritional quality traits. Int J Genet 9(5):278–283
Gantait S, Gunri SK, Kundu R, Chatterjee S (2017) Evaluation of genetic divergence in Spanish bunch groundnut (Arachis hypogaea Linn.) genotypes. Plant Breeding and Biotechnology,. 5:163–1713
Gautami B, Fonce D, Pandey MK, Moretzsohn MC, Sujay V, Quin H, Hong Y, Faye I, Chen X, Prakash AB, Shah T, M,Gowda MVC, Nigam SN, Liang X, Hoisington DA, Guo B, Bertioli DJ, Rami JF, Varshney RK (2012) An International reference consensus genetic map with 897 marker loci based on 11 mapping populations for tetraploid groundnut (Arachishypogeae L.). PLoS ONE 7(7):1–11
Gupta RP, Vachhani JH (2015) Genetic divergence studies in Virginia groundnut (ArachishypogaeaL.). Trend Biosci 8:543–547
He G, Meng R, Newman M, Gao G, Pittman RN, Prakash CS (2003) Microsatellites as DNA markers in cultivated peanut (Arachis hypogaea L.). BMC Plant Biol 3(1):1–6
Hopkins M, Casa A, Wang T, Mitchell S, Dean R, Kochert G, Kresovich S (1999) Discovery and characterization of polymorphic simple sequence repeats (SSRs) in peanut. Crop science, 39(4): 1243–1247
Krishna GK, Zhang J, Burow M, Pittman RN, Delikostadinov SG, Lu Y, Puppala N (2004) Genetic diversity analysis in Valencia peanut (Arachishypogaea L.) using microsatellite markers, Cellular & Molecular Biology Letters, (9):685–697
Liu K, Muse M (2005) Power Marker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Mahalanobis PC (1936) On the generalized distance in statistics. Proceedings of the National Institute of Sciences (Calcutta) 2: 49–55
Mahesh RH, Hasan K (2018) Analysis of genetic diversity of groundnut (Arachis hypogaea L.) genotypes collected from various parts of India. J Pharmacognosy Phytochem 7(2):1100–1103
Murty B, Arunachalam V (1966) The nature of genetic divergence in relation to breeding system in some crop plants. Indian J Genet 26:188–198
Nei M (1990) Heterozygosity and genetic-distance - a citation classic commentary on estimation of average heterozygosity and genetic-distance from a small number of individuals. Genetics 89:583–590
Oteng-Frimpong R, Sriswathi M, Ntare BR, Dakor F (2015) Assessing the genetic diversity of 48 groundnut (Arachis hypogaea L.) genotypes in the Guinea savanna agro-ecology of Ghana, using microsatellite-based markers. Afr J Biotechnol 14(32):2485–2485
Raghuwanshi SS, Kachhadia V, Vachhani J, Jivani L, Patel M (2016) Genetic divergence in ground nut (Arachis hypogaea L.). Electron J Plant Breed 7(1):145–151
Rao B, Murty VAR, Subramanyam RJ (1952) The amylase and amylopectin content of rice and their influence on cooking quality of cereals. Proc. Indian Acad. Sci, 36: 70–74
Raza M, Sekhar MR, Priya MS, Prasad T, Rajarajeswari V (2018) Genetic diversity studies for pod yield and nutrient uptake related traits in groundnut. Indian J Agricultural Res 52(4):380–385
Saleem M, Naidu G, Nadaf H, Tippannavar P (2017) Genetic diversity in groundnut (Arachis hypogaea L.) based on reaction to biotic stresses and productivity parameters, Legume Research, Online ISSN:0976 – 0571
Salem KF, Sallam A (2016) Analysis of population structure and genetic diversity of Egyptian and exotic rice (Oryza sativa L.) genotypes. Comptes rendus biologies, 339(1): 1–9
Singh S, Singh A, Kalpana S, Misra S (2010) Genetic diversity for growth, yield and quality traits in groundnut (Arachis hypogaea L.). Indian J Plant Physiol 15(3):267–271
Author Information
Department of Oilseeds, Centre for Plant Breeding and Genetics, Tamil Nadu Agricultural University, Tamil Nadu, India