HPTLC quantification of quercetin from leaf extracts of C. occidentalis L. and its inhibitory activity against protease enzyme of P. falciparum
Research Articles | Published: 11 June, 2024
First Page: 1804
Last Page: 1816
Views: 1780
Keywords: HPTLC, Quercetin, Inhibitory activity, Plasmepsin-II, Molecular docking, MD simulation
Abstract
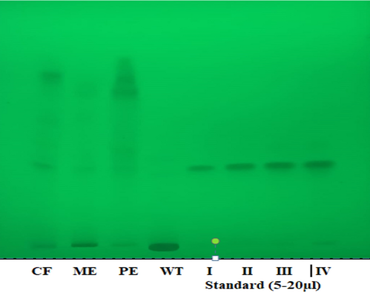
Most of the therapeutically relevant antimalarial medications target the parasite during the blood stage of its life cycle. As a result, new antimalarial medicines have focused on inhibiting the breakdown of host-cell hemoglobin. C. occidentalis was reported to have antimalarial activity and proved very effective in the conventional Indian medical system. This study aimed to analyze the quercetin content of four different leaf extracts of C. occidentalis using a validated HPTLC method and to evaluate the inhibitory activity of quercetin against Plasmepsin-II of P. falciparum using in silico approach. HPTLC results revealed that the CF extract of C. occidentalis had the highest quercetin content with 4.10% (w/w), followed by the ME extract with 2.37% (w/w), PE extract with 2.09% (w/w), and WT extract with 1.79% (w/w). In silico investigations of quercetin with the Plasmepsin-II enzyme of P. falciparum portrayed excellent binding affinities of (− 7.5 kcal/mol) and convenient molecular interactions with important amino acid residues of the target receptor as compared with control. Furthermore, MD simulation study confirmed the finding of molecular docking by depicting the stability of the simulated system both in water and in vacuum. Finally, the study suggested that the chloroform extract of C. occidentalis is the most suitable and efficient solvent for extracting high-quantity and quality quercetin. The findings of the molecular docking and MD simulation analysis further supported the potential inhibitory activity of quercetin against Plasmepsin-II. This research contributes to the understanding of quercetin and its potential applications in combating antimalarial drug resistance.
References
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1:19–25
Adewole KE (2020) Nigerian antimalarial plants and their anticancer potential: a review. J Integr Med 18(2):92–113. https://doi.org/10.1016/j.joim.2020.01.001
Alotaibi BS, Ijaz M, Buabeid M, Kharaba ZJ, Yaseen HS, Murtaza G (2021) Therapeutic effects and safe uses of plant-derived polyphenolic compounds in cardiovascular diseases: a review. Drug Des Develop Ther 2021:4713–4732
Asojo OA, Gulnik SV, Afonina E, Yu B, Ellman JA, Haque TS, Silva AM (2003) Novel uncomplexed and complexed structures of plasmepsin II, an aspartic proteasefrom Plasmodium falciparum. J Mol Biol 327(1):173–181. https://doi.org/10.1016/S0022-2836(03)00036-6
Berendsen HJ, Postma JV, Van Gunsteren WF, DiNola ARHJ, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81(8):3684–3690
Bernetti M, Bussi G (2020) Pressure control using stochastic cell rescaling. J Chem Phys 153(11):1
Bhardwaj VK, Purohit R (2020) A new insight into protein–protein interactions and the effect of conformational alterations in PCNA. Int J Biol Macromol 148:999–1009. https://doi.org/10.1016/j.ijbiomac.2020.01.212
Brooks BR, Brooks CL III, Mackerell AD Jr, Nilsson L, Petrella RJ, Roux B, Karplus M (2009) CHARMM: the biomolecular simulation program. J Comput Chem 30(10):1545–1614
Bylicka-Szczepanowska E, Korzeniewski K (2022) Asymptomatic malaria infections in the time of COVID-19 pandemic: experience from the Central African Republic. Int J Environ Res Public Health 19(6):3544
Cheng J, Hao Y, Shi Q, Hou G, Wang Y, Wang Y, Yu G (2023) Discovery of novel Chinese medicine compounds targeting 3CL protease by virtual screening and molecular dynamics simulation. Molecules 28(3):937
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7(January):1–13. https://doi.org/10.1038/srep42717
De La Nuez A, Rodríguez R (2008) Current methodology for the assessment of ADME-Tox properties on drug candidate molecules. Biotecnol Apl 25(2):97–110
DeLano WL (2002) The PyMOL molecular graphics system. http://www.pymol.org/
Discovery Studio Visualization (2016) (Internet) (Cited 2 Dec 2016). http://accelrys.com/products/collaborativescience/biovia-discovery-studio/visualizationdownload.php
ICH Harmonised Tripartite Guideline (2005) ICH harmonised tripartite guideline validation of analytical procedures
Hess B, Bekker H, Berendsen HJ, Fraaije JG (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18(12):1463–1472
Muhseen ZT, Hameed AR, Al-Bhadly O, Ahmad S, Li G (2021) Natural products for treatment of Plasmodium falciparum malaria: an integrated computational approach. Comput Biol Med 134(March):104415. https://doi.org/10.1016/j.compbiomed.2021.104415
Pillay P, Maharaj VJ, Smith PJ (2008) Investigating South African plants as a source of new antimalarial drugs. J Ethnopharmacol 119(3):438–454. https://doi.org/10.1016/j.jep.2008.07.003
Pires DEV, Blundell TL, Ascher DB (2015) pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem 58(9):4066–4072. https://doi.org/10.1021/acs.jmedchem.5b00104
Purohit R (2014) Role of ELA region in auto-activation of mutant KIT receptor: a molecular dynamics simulation insight. J Biomol Struct Dyn 32(7):1033–1046. https://doi.org/10.1080/07391102.2013.803264
Shabir I, Kumar Pandey V, Shams R, Dar AH, Dash KK, Khan SA, Pandiselvam R (2022) Promising bioactive properties of quercetin for potential food applications and health benefits: a review. Front Nutr 9:999752
Shibeshi MA, Kifle ZD, Atnafie SA (2020) Antimalarial drug resistance and novel targets for antimalarial drug discovery. Infect Drug Resist 13:4047–4060. https://doi.org/10.2147/IDR.S279433
Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Mackerell AD Jr (2010) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 31(4):671–690
Verma AK, Bala HA, Muhammad II, Muhammad A, Kori AR, Barik M (2020) Virtual screening, molecular docking, pharmacokinetic, physicochemical and medicinal properties of potential curcumin derivatives against SARS-CoV-2 main protease (Mpro). Asian J Pharm Anal Med Chem 8(4):153–179
Verma AK, Srivastava VK, Srivastava SK (2023) Identification of corticosteroids as potential inhibitor against glycolytic enzyme hexokinase II role in cancer glycolysis pathway: a molecular docking study. Vegetos. https://doi.org/10.1007/s42535-022-00564-3
Author Information
School of Bioengineering and Bioscience, Lovely Professional University, Phagwara, India