Identification and characterization of genes responsive to drought and heat stress in rice (Oryza sativa L.)
Research Articles | Published: 30 March, 2021
First Page: 309
Last Page: 317
Views: 4016
Keywords: Rice, Microarray, Heat, Drought, DEGs
Abstract
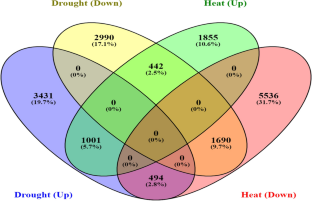
Rice is considered as a major source of food for most people worldwide. Global rice production is under threat as a result of adverse effects of heat and drought stress. It is therefore necessary to safeguard its sustainable production. This study was carried out to characterize differentially expressed genes (DEGs) under both drought and heat stresses in rice. 1001 DEGs were determined to show up-regulation under heat and drought stresses, while 1690 DEGs were commonly down-regulated. Functional classification analysis generated 22 and 37 related gene groups from the commonly up-regulated and down-regulated genes respectively. Functional characterization revealed 38.1% and 45.2% DEGs annotating for Biological Process, 53.2% and 59.2% DEGs were for Cellular Components and 54.8% and 52.4% DEGs were for Molecular Function in the up and down commonly regulated DEGs respectively. KEGG analysis demonstrated that most of the up-regulated DEGs were particularly enriched in metabolic pathways and in the biosynthesis of secondary metabolites, while the down regulated genes were mostly enriched in pathways involving ribosomes, and in purine and pyrimidine metabolisms. These results could be helpful in further analysis and understanding of heat and drought stress tolerance in rice.
References
- Ashburner M (2000) A biologist’s view of the Drosophila genome annotation assessment project. Genome Res 10(4):391–393
- Barozai MYK, Husnain T (2011) Identification of biotic and abiotic stress up-regulated ESTs in Gossypiumarboreum. Mol Biol Rep 39(2):1011–1018
- Barozai MYK, Wahid HA (2012) In silico identification and characterization of cumulative abiotic stress responding genes in Potato (Solanumtuberosum L.). Pak J Bot 44:57–69
- CGIAR (2016) The global staple. http://ricepedia.org/rice-asfood/the-global-staple-rice-consumers. Accessed 15 Sept 2020
- Cohen J (1960) A coefficient of agreement for nominal scales. Educ Psychol Meas 20(1):37–46
- Eisen M, Spellman P, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95(25):14863–14868
- Fitzgerald MA, Resurreccion AP (2009) Maintaining the yield of edible rice in a warming world. Funct Plant Biol 36:1037–1045
- Gao WR, Wang XS, Liu QY, Peng H, Chen C, Li JG, Ma H (2008) Comparative analysis of ESTs in response to drought stress in chickpea (C. arietinum L.). BBRC 376(3):578–583
- González-Schain N, Dreni L, Lawas LMF et al (2016) Genome-wide transcriptome analysis during anthesis reveals new insights into the molecular basis of heat stress responses in tolerant and sensitive rice varieties. Plant Cell Physiol 57:57–68
- Hanif S, Saleem MF, Sarwar M et al (2020) Biochemically triggered heat and drought stress tolerance in rice by proline application. J Plant Growth Regul 40:305–315
- IPCC (2014) Climate change synthesis report. In: Team CW, Pachauri RK, Meyer LA (eds) Contribution of working groups I, II and III to the fifth assessment report of the intergovernmental panel on climate change. Cambridge University Press, Geneva
- Jagadish SVK, Craufurd PQ, Wheeler TR (2008) Phenotyping parents of mapping populations of rice for heat tolerance during anthesis. Crop Sci 48:1140–1146
- Jagadish SVK, Muthurajan R, Rang ZW et al (2011) Spikelet proteomic response to combined water deficit and heat stress in rice (Oryzasativa cv. N22). Rice 4:1–11
- Kadam NN, Struik PC, Rebolledo MC et al (2018) Genome-wide association reveals novel genomic loci controlling rice grain yield and its component traits under water-deficit stress during the reproductive stage. J Exp Bot 69:4017–4032
- Kazuo N, Yusuke I, Shinozaki KY (2009) Transcriptional regulatory networks in response to abiotic stresses in arabidopsis and grasses. Plant Physiol 149:88–95
- Kumar A, Verulkar S, Dixit S et al (2009) Yield and yield-attributing traits of rice (Oryzasativa L.) under lowland drought and suitability of early vigor as a selection criterion. Field Crop Res 114:99–107
- Lata C, Bhutty S, Bahadur RP, Majee M, Prasad M (2011) Association of an SNP in a novel DREB2-like gene SiDREB2 with stress tolerance in foxtail millet [Setariaitalica (L.)]. J Exp Bot 62(10):3387–3401
- Lawas LMF, Shi W, Yoshimoto M et al (2018) Combined drought and heat stress impact during flowering and grain filling in contrasting rice cultivars grown under field conditions. Field Crops Res 229:66–77
- Li X, Lawas LMF, Malo R et al (2015) Metabolic and transcriptomic signatures of rice floral organs reveal sugar starvation as a factor in reproductive failure under heat and drought stress. Plant Cell Environ 38:2171–2192
- Lorković ZJ (2009) Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci 14(4):229–236
- Mahalingam R (2017) Phenotypic, physiological and malt quality analyses of US barley varieties subjected to short periods of heat and drought stress. J Cereal Sci 76:199–205
- Mahrookashani A, Siebert S, Hüging H et al (2017) Independent and combined effects of high temperature and drought stress around anthesis on wheat. J Agron Crop Sci 203:453–463
- Mittler R, Finka A, Goloubinoff P (2012) How do plants feel the heat? Trends Biochem Sci 37(3):118–125
- Morishita T, Kojima Y, Maruta T, Nishizawa-Yokoi A, Yabuta Y, Shigeoka S (2009) Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant Cell Physiol 50(12):2210–2222
- Mukamuhirwa A, Persson Hovmalm H, Bolinsson H, Ortiz R, Nyamangyoku O, Johansson E (2019) Concurrent drought and temperature stress in rice—a possible result of the predicted climate change: effects on yield attributes, eating characteristics, and health promoting compounds. Int J Environ Res Public Health 16(6):1043
- Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2012) NAC transcription factors in plant abiotic stress responses. BBA Gene Regul Mech 1819(2):97–103
- Niinemets Ü (2016) Uncovering the hidden facets of drought stress: secondary metabolites make the difference. Tree Physiol 36(2):129–132
- Nivedita Yadav PK, Gautam B (2015) Gene expression profiling of transcription factors of arabidopsis thaliana using microarray data analysis. IJARCSSE 5(4):783–793
- Pantuwan G, Fukai S, Cooper M et al (2002) Yield response of rice (Oryzasativa L.) genotypes to different types of drought under rainfed lowlands Part 1. Grain yield and yield components. Field Crops Res 73:153–168
- Perdomo JA, Conesa M, Medrano H et al (2015) Effects of long-term individual and combined water and temperature stress on the growth of rice, wheat and maize: relationship with morphological and physiological acclimation. Physiol Plant 155:149–165
- Pinoli P, Chicco D, Masseroli M (2015) Computational algorithms to predict gene ontology annotations. BMC Bioinform 16(6):S4
- Pradhan GP, Prasad PVV, Fritz AK et al (2012) Effects of drought and high temperature stress on synthetic hexaploid wheat. Funct Plant Biol 39:190–198
- Prasad PVV, Pisipati SR, Momcilovic I, Ristic Z (2011) Independent and combined effects of high temperature and drought stress during grain filling on plant yield and chloroplast EF-Tu expression in spring wheat. J Agron Crop Sci 197:430–441
- Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Monitoring expression profiles of rice genes under cold, drought, and highsalinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133:1755–1767
- Rang ZW, Jagadish SVK, Zhou QM et al (2011) Effect of high temperature and water stress on pollen germination and spikelet fertility in rice. Environ Exp Bot 70:58–65
- Rizhsky L, Liang H, Mittler R (2002) The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol 130:1143–1151
- Rizhsky L, Liang H, Shuman J et al (2004) When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134:1683–1696
- Sasidharan R, Mustroph A, Boonman A, Akman M, Ammerlaan AMH, Breit T, Schranz ME, Voesenek LACJ, Van Tienderen PH (2013) Root transcript profiling of two Rorippa species reveals gene clusters associated with extreme submergence tolerance. Plant Physiol 163:1277–1292
- Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T et al (2002) Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high salinity stresses using a full length cDNA microarray. Plant J 31:279–292
- Shaar-Moshe L, Blumwald E, Peleg Z (2017) Unique physiological and transcriptional shifts under combinations of salinity, drought, and heat. Plant Physiol 174:421–434
- Shi W, Lawas LMF, Raju BR et al (2016) Acquired thermo-tolerance and trans-generational heat stress response at flowering in rice. J Agron Crop Sci 202:309–319
- Suzuki N, Rivero RM, Shulaev V et al (2014) Abiotic and biotic stress combinations. New Phytol 203:32–43
- Teixeira EI, Fischer G, Velthuizen HV, Walter C, Ewert F (2013) Global hot-spots of heat stress on agricultural crops due to climate change. Agric For Meteorol 170:206–215
- Torres RO, McNally KL, Cruz CV et al (2013) Screening of rice Genebank germplasm for yield and selection of new drought tolerance donors. Field Crops Res 147:12–22
- Trachsel S, Leyva M, Lopez M et al (2016) Identification of tropical maize germplasm with tolerance to drought, nitrogen deficiency, and combined heat and drought stresses. Crop Sci 56:3031–3045
- Tula S, Ansari MW, Babu AP, Pushpalatha G, Sreenu K, Sarla N et al (2013) Physiological assessment and allele mining in rice cultivars for salinity and drought stress tolerance. Vegetos 26(1):00–00
- Wahid A, Gelani S, Ashraf M, Foolad MR (2007) Heat tolerance in plants: an overview. Environ Exp Bot 61(3):199–223
- Wassmann R, Jagadish SVK, Sumfleth K et al (2009) Regional vulnerability of climate change impacts on Asian rice production and scope for adaptation. In: Sparks DL (ed) Advances in agronomy. Academic Press, Burlington, pp 91–133
- Zandalinas SI, Rivero RM, Martínez V, Gómez-Cadenas A, Arbona V (2016) Tolerance of citrus plants to the combination of high temperatures and drought is associated to the increase in transpiration modulated by a reduction in abscisic acid levels. BMC Plant Biol 16:105
- Zhao F, Zhang D, Zhao Y et al (2016) The difference of physiological and proteomic changes in maize leaves adaptation to drought, heat, and combined both stresses. Front Plant Sci 7:1471
- Zhou J, Wang X, Jiao Y, Qin Y, Liu X, He K, Chen C, Ma L, Wang J, Xiong L, Zhang Q, Fan L, Deng XW (2007) Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Mol Biol 63(5):591–608
Author Information
Indian Agricultural Research Institute, New Delhi, India