Identification of high yielding and yellow mosaic virus resistance blackgram (Vigna mungo L. Wilczek) genotypes using multivariate analysis
Singh Varsha, Singh Mahendra Narain, Singh Anil Kumar, Madankar Kartik, Sinha B.
Research Articles | Published: 30 March, 2023
First Page: 363
Last Page: 372
Views: 3863
Keywords: Blackgram, Genetic variability, MYMV resistance, PDI, Principal component
Abstract
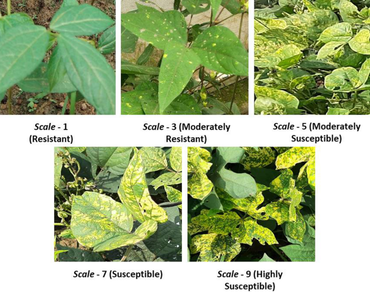
21 blackgram genotypes, including the infector ‘Co 5’, were raised in a Completely Randomized Block design with two replications for two consecutive years (2018 and 2019) to screen blackgram yellow mosaic virus (MYMV) disease resistance under field conditions. Two genotypes viz., NDU 1 and Co 5 were highly susceptible to MYMV having PDI > 40% while other genotypes were resistant with PDI up to 10%. In general, genotypes with higher PDI (> 40%) had a lower number of pods per plant (NPP) and vice versa. The average number of susceptible genotypes pods per plant showed 23.75% reduction in number of pods per plant compared with resistant genotypes. These genotypes have undergone multivariate analysis including genetic variability, correlation, divergence and principal component analysis for eleven biometric traits. Principal component analysis revealed significant variation among traits with the first three principal components explaining approximately 74% of the total variation. Seed yield per plant; days to 50% flowering and percent disease index are important traits determining most of the variation. Mash 338, R3/12, R3/28 and PU 31 are the promising blackgram genotypes identified in the present study. These genotypes will be used to develop high yielding MYMV resistant blackgram varieties having other desirable yield traits.
References
Anderson TW (1972) An introduction to multivariate statistical analysis. Wiley Eastern Private Limited, New Delhi, p 512
Iqbal U, Iqbal SM, Afzal R, Jamal A, Farooq MA et al (2011) Screening of mungbean germplasm against mungbean yellow mosaic virus (MYMV) under field conditions. Pak J Phytopathol 23:48–51
Jeberson MS, Shashidhar KS, Singh AK (2019) Genetic variability, principal component and cluster analyses in black gram under Foot-hills conditions of Manipur. Legum Res 42(4):454–460
Kaufman L, Rousseeuw PJ (1990) Finding groups in data: an introduction to cluster analysis. Wiley, New York
Konda CR, Salimath PM, Mishra MN (2009) Genetic variability studies for productivity and its components in blackgram (Vigna munga L. Hepper). Legum Res 32:59–61
Korattukudy CA, Tamilarasi AP, Rengasamy K, Devadhasan S, Shunmugavel S, Yasin JK, Madhavan AP (2022) Genetic diversity analysis in blackgram (Vigna mungo) genotypes using microsatellite markers for resistance to Yellow mosaic virus. Plant Prot Sci 58:110–124
Kumar VG, Vanaja M, Sathish P, Vagheera P, Shishodia SS, Razak A (2014) Correlation and path analysis seed yield and yield contributing components of black gram (Vigna mungo L. Hepper) under rainfed condition from Andhra Pradesh, India. Int J Appl Biol Pharm Technol 5:137–140
Kumari S, Sahni S, Prasad BD (2020) Screening of urdbean germplasm for high seed yield coupled with resistance to mungbean yellow mosaic virus (MYMV). Curr J Appl Sci Technol 39(1):100–106
Lavanya GR, Srivastava J, Ranade SA (2008) Molecular assessment of genetic diversity in mung bean germplasm. J Genet 87:65–74
Palanivelu S, Narayana M, Palaniappan V, Natarajan G, Gandhi K (2021) Screening for urdbean leaf crinkle disease at feld condition in blackgram [Vigna mungo (L.) Hepper]. Vegetos. https://doi.org/10.1007/s42535-019-00057-w
Panigrahi KK, Mohanty A, Baisakh B (2014) Genetic divergence, variability and character association in landraces of blackgram (Vigna mungo L. Hepper) from Odisha. J Crop Weed 10:155–165
Pavishna M, Kannan R, Arumugam Pillai M, Rajinimala N (2019) Screening of blackgram genotypes against mung bean yellow mosaic virus disease. J Pharmacogn Phytochem 8(3):4313–4318
Popat R, Patel R, Parmar D (2020) Variability: genetic variability analysis for plant breeding research. R package version 0.1.0.
RStudio Team (2020) RStudio: integrated development for R. RStudio PBC, Boston
Sarkar M, Kundaram S (2016) Multivariate analysis in some genotypes of blackgram [Vigna radiata (L.) Wilczek] on the basis of agronomic traits of two consecutive growing cycles. Legume Res 39:1–5
Singh G, Sharma YR, Kaur L (1992) Method of rating blackgram yellow mosaic virus in blackgram and blackgram. Plant Dis Res 7:1–6
Singh V, Singh P, Dheer M, Nath S, Punia SS (2017) Assessment of genetic diversity in chickpea (Cicer arietinum L) Germplasm. J Plant Dev Sci 9(7):699–702
Singh V, Singh P, Kumar A, Shiva N (2018) Estimation of genetic variability parameters in chickpea (Cicer arietinum L) germplasm. J Pharmacogn Phytochem 7(2):1204–1206
Singh L, Kumar A, Kaur S, Gill RK (2020) Multivariate analysis of yield contributory traits for selection criteria in Blackgram (Vigna mungo L. Hepper). Electron J Plant Breed 11(4):1134–1142
Singh SV, Singh AK, Singh MN, Sinha B (2022) Comparative study of crossability behavior in intra-specific and inter-specific crosses of Vigna radiata and Vigna mungo. Legum Res. https://doi.org/10.18805/LR-5002
Author Information
Department of Genetics and Plant Breeding, Institute of Agriculture, Banaras Hindu University, Varanasi, India