Plant-derived natural compounds aiding SOCS1 mediated JAK1 inhibition, a novel mechanism of combinatorial cancer chemotherapy
Garg Saksham, Kumar Sunil, Anand Ashutosh, Menon Tarunya, Sharma Nikita, Singh Japneet, Chawla Siddharth, Das Asmita, Kumar Sunil, Anand Ashutosh
Research Articles | Published: 12 February, 2022
First Page: 707
Last Page: 722
Views: 4875
Keywords: SOCS1, JAK1, Ubiquitination, STAT, HNSC, Structural-based drug discovery, Combinatorial therapy
Abstract
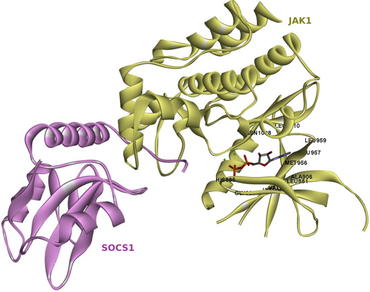
Numerous drugs have been used in the past to treat HNSC cancer through tumor suppression and immune modulation mechanisms. However, none of them achieved complete tumor remission. Synthetic drugs targeting tumor cells have side effects, and the tumor often acquires resistance against them. A subfamily of tyrosine kinases called Janus Kinases (JAKs) is observed to be over-expressed in various solid tumors, including HNSC. JAKs directly activate a family of transcription factors, Signal Transducers and Activators of Transcription (STATs) and induce a signaling cascade collectively known as JAK/STAT pathways. STATs are responsible for the regulated production of many inflammatory cytokines and growth factors that are beneficial to the tumor cells, favouring them to sustain themselves in a hostile microenvironment. Hence, inhibitors of JAK have been explored previously and SOCS 1 has been shown to be a known direct and most potent inhibitor of JAK1 among the family of SOCSs proteins. The study presented here proposes a mechanism to inhibit the JAK/STAT pathway by inhibiting the JAK1 protein using small molecules of plant origin. The study thereby proposes three inhibitors viz., withaferin A, silymarin, and hypericin, to have significant potential to inhibit JAK1 protein, known to be upregulated in tumors. SOCS1 was also identified to be upregulated in an HNSC tumor samples and is known to inhibit JAK-STAT pathway. Our 3 potent inhibitors, withaferin A, silymarin, and hypericin had the ability to also bind to the SOCS1-JAK1complex thus stabilizing it thus further potentiating the inhibition of JAK-STAT pathway. The three inhibitors explored in the present study can prevent JAK phosphorylation and activation in preventive and therapeutic application. The study proposes a therapy that can be employed in combination with other cancer therapies, thus increasing the overall efficiency of the treatment.
References
Lawrence MS, Sougnez C, Lichtenstein L, Cibulskis K, Lander E, Gabriel SB, Getz G, Ally A, Balasundaram M, Birol I, Bowlby R, Brooks D, Butterfield YSN, Carlsen R, Cheng D, Chu A, Dhalla N, Guin R, Holt RA, Jones SJM, Lee D, Li HI, Marra MA, Mayo M, Moore RA, Mungall AJ, Robertson AG, Schein JE, Sipahimalani P, Tam A, Thiessen N, Wong T, Protopopov A, Santoso N, Lee S, Parfenov M, Zhang J, Mahadeshwar HS, Tang J, Ren X, Seth S, Haseley P, Zeng D, Yang L, Xu AW, Song X, Pantazi A, Bristow CA, Hadjipanayis A, Seidman J, Chin L, Park PJ, Kucherlapati R, Akbani R, Casasent T, Liu W, Lu Y, Mills G, Motter T, Weinstein J, Diao L, Wang J, Hong Fan Y, Liu J, Wang K, Auman JT, Balu S, Bodenheimer T, Buda E, Hayes DN, Hoadley KA, Hoyle AP, Jefferys SR, Jones CD, Kimes PK, Liu Y, Marron JS, Meng S, Mieczkowski PA, Mose LE, Parker JS, Perou CM, Prins JF, Roach J, Shi Y, Simons JV, Singh D, Soloway MG, Tan D, Veluvolu U, Walter V, Waring S, Wilkerson MD, Wu J, Zhao N, Cherniack AD, Hammerman PS, Tward AD, Pedamallu CS, Saksena G, Jung J, Ojesina AI, Carter SL, Zack TI, Schumacher SE, Beroukhim R, Freeman SS, Meyerson M, Cho J, Noble MS, DiCara D, Zhang H, Heiman DI, Gehlenborg N, Voet D, Lin P, Frazer S, Stojanov P, Liu Y, Zou L, Kim J, Muzny D, Doddapaneni HV, Kovar C, Reid J, Morton D, Han Y, Hale W, Chao H, Chang K, Drummond JA, Gibbs RA, Kakkar N, Wheeler D, Xi L, Ciriello G, Ladanyi M, Lee W, Ramirez R, Sander C, Shen R, Sinha R, Weinhold N, Taylor BS, Aksoy BA, Dresdner G, Gao J, Gross B, Jacobsen A, Reva B, Schultz N, Sumer SO, Sun Y, Chan TA, Morris LG, Stuart J, Benz S, Ng S, Benz C, Yau C, Baylin SB, Cope L, Danilova L, Herman JG, Bootwalla M, Maglinte DT, Laird PW, Triche T, Weisenberger DJ, Van Den Berg DJ, Agrawal N, Bishop J, Boutros PC, Bruce JP, Byers LA, Califano J, Carey TE, Chen Z, Cheng H, Chiosea SI, Cohen E, Diergaarde B, Egloff AM, El-Naggar AK, Ferris RL, Frederick MJ, Grandis JR, Guo Y, Haddad RI, Harris T, Hui ABY, Lee JJ, Lippman SM, Liu FF, McHugh JB, Myers J, Ng PKS, Perez-Ordonez B, Pickering CR, Prystowsky M, Romkes M, Saleh AD, Sartor MA, Seethala R, Seiwert TY, Si H, Van Waes C, Waggott DM, Wiznerowicz M, Yarbrough WG, Zhang J, Zuo Z, Burnett K, Crain D, Gardner J, Lau K, Mallery D, Morris S, Paulauskis J, Penny R, Shelton C, Shelton T, Sherman M, Yena P, Black AD, Bowen J, Frick J, Gastier-Foster JM, Harper HA, Leraas K, Lichtenberg TM, Ramirez NC, Wise L, Zmuda E, Baboud J, Jensen MA, Kahn AB, Pihl TD, Pot DA, Srinivasan D, Walton JS, Wan Y, Burton RA, Davidsen T, Demchok JA, Eley G, Ferguson ML, Mills Shaw KR, Ozenberger BA, Sheth M, Sofia HJ, Tarnuzzer R, Wang Z, Yang L, Zenklusen JC, Saller C, Tarvin K, Chen C, Bollag R, Weinberger P, Golusiński W, Golusiński P, Ibbs M, Korski K, Mackiewicz A, Suchorska W, Szybiak B, Curley E, Beard C, Mitchell C, Sandusky G, Ahn J, Khan Z, Irish J, Waldron J, William WN, Egea S, Gomez-Fernandez C, Herbert L, Bradford CR, Chepeha DB, Haddad AS, Jones TR, Komarck CM, Malakh M, Moyer JS, Nguyen A, Peterson LA, Prince ME, Rozek LS, Taylor EG, Walline HM, Wolf GT, Boice L, Chera BS, Funkhouser WK, Gulley ML, Hackman TG, Hayward MC, Huang M, Rathmell WK, Salazar AH, Shockley WW, Shores CG, Thorne L, Weissler MC, Wrenn S, Zanation AM, Brown BT, Pham M (2015) Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517:576–582. https://doi.org/10.1038/nature14129
Ahn R, Sabourin V, Bolt AM, Hébert S, Totten S, De Jay N, Festa MC, Young YK, Im YK, Pawson T, Koromilas AE, Muller WJ, Mann KK, Kleinman CL (2017) Ursini-Siegel, the Shc1 adaptor simultaneously balances Stat1 and Stat3 activity to promote breast cancer immune suppression. Nat Commun 81:1–14. https://doi.org/10.1038/ncomms14638
Akinleye A, Rasool Z (2019) Immune checkpoint inhibitors of PD-L1 as cancer therapeutics. J Hematol Oncol 12:92. https://doi.org/10.1186/s13045-019-0779-5
Babon JJ, Sabo JK, Soetopo A, Yao S, Bailey MF, Zhang J-G, Nicola NA, Norton RS (2008) The SOCS box domain of SOCS3: structure and interaction with the ElonginBC-Cullin5 ubiquitin ligase. J Mol Biol 381:928–940. https://doi.org/10.1016/j.jmb.2008.06.038
Berman HM (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242. https://doi.org/10.1093/nar/28.1.235
Bowers KJ, Chow DE, Xu H, Dror RO, Eastwood MP, Gregersen BA, Klepeis JL, Kolossvary I, Moraes MA, Sacerdoti FD, Salmon JK, Shan Y, Shaw DE (2006) Scalable algorithms for molecular dynamics simulations on commodity clusters. In: ACM/IEEE SC IEEE, pp 43–43. https://doi.org/10.1109/SC.2006.54
Bradner JE, Hnisz D, Young RA (2017) Transcriptional addiction in cancer. Cell 168:629–643. https://doi.org/10.1016/j.cell.2016.12.013
Chikuma S, Kanamori M, Mise-Omata S, Yoshimura A (2017) Suppressors of cytokine signaling: Potential immune checkpoint molecules for cancer immunotherapy. Cancer Sci 108:574–580. https://doi.org/10.1111/cas.13194
Cho KH, Jeong KJ, Shin SC, Kang J, Park CG, Lee HY (2013) STAT3 mediates TGF-β1-induced TWIST1 expression and prostate cancer invasion. Cancer Lett 336:167–173. https://doi.org/10.1016/j.canlet.2013.04.024
Choi H, Cho SY, Pak HJ, Kim Y, Choi J, Lee YJ, Gong BH, Kang YS, Han T, Choi G, Cho Y, Lee S, Ryoo D, Park H (2017) NPCARE: database of natural products and fractional extracts for cancer regulation. J Chem Inform 9:2. https://doi.org/10.1186/s13321-016-0188-5
Colavito SA (2020) AXL as a target in breast cancer therapy. J Oncol. https://doi.org/10.1155/2020/5291952
Demaria M, Giorgi C, Lebiedzinska M, Esposito G, D’Angeli L, Bartoli A, Gough DJ, Turkson J, Levy DE, Watson CJ, Wieckowski MR, Provero P, Pinton P, Poli V (2010) A STAT3-mediated metabolic switch is involved in tumour transformation and STAT3 addiction. Aging (Albany NY) 2:823–842. https://doi.org/10.18632/aging.100232
Gamero AM, Young MR, Mentor-Marcel R, Bobe G, Scarzello AJ, Wise J, Colburn NH (2010) STAT2 contributes to promotion of colorectal and skin carcinogenesis. Cancer Prev Res (Phila) 3:495. https://doi.org/10.1158/1940-6207.CAPR-09-0105
Goel S, DeCristo MJ, Watt AC, BrinJones H, Sceneay J, Li BB, Khan N, Ubellacker JM, Xie S, Metzger-Filho O, Hoog J, Ellis MJ, Ma CX, Ramm S, Krop IE, Winer EP, Roberts TM, Kim H-J, McAllister SS, Zhao JJ (2017) CDK4/6 inhibition triggers anti-tumour immunity. Nature 548:471–475. https://doi.org/10.1038/nature23465
Goel RK, Singh D, Lagunin A, Poroikov V (2011) PASS-assisted exploration of new therapeutic potential of natural products. Med Chem Res 20:1509–1514. https://doi.org/10.1007/s00044-010-9398-y
Han Y, Liu D, Li L (2020) PD-1/PD-L1 pathway: current researches in cancer. Am J Cancer Res 10:727–742. http://www.ncbi.nlm.nih.gov/pubmed/32266087
Harjunpää H, Guillerey C (2020) TIGIT as an emerging immune checkpoint. Clin Exp Immunol 200:108–119. https://doi.org/10.1111/cei.13407
Hou X, Zhou G, Fan Y, Zhang Q, Xiang C, Cao F, Yao S (2020) The high expression of CD276/HAVCR2 and CD163 is an adverse immune subtype of glioblastoma and is closely related to epithelial-mesenchymal transition. https://doi.org/10.21203/rs.3.rs-31174/v1
Hu Y, Sun H, Hu J, Zhang X (2020) LncRNA DLX6-AS1 promotes the progression of neuroblastoma by activating STAT2 via targeting miR-506-3p. Cancer Manag Res 12:7451. https://doi.org/10.2147/CMAR.S252521
Huang Q, Duan L, Qian X, Fan J, Lv Z, Zhang X, Han J, Wu F, Guo M, Hu G, Du J, Chen C, Jin Y (2016) IL-17 promotes angiogenic factors IL-6, IL-8, and Vegf production via Stat1 in lung adenocarcinoma. Sci Rep 6:36551. https://doi.org/10.1038/srep36551
Huang Y-H, Zhu C, Kondo Y, Anderson AC, Gandhi A, Russell A, Dougan SK, Petersen B-S, Melum E, Pertel T, Clayton KL, Raab M, Chen Q, Beauchemin N, Yazaki PJ, Pyzik M, Ostrowski MA, Glickman JN, Rudd CE, Ploegh HL, Franke A, Petsko GA, Kuchroo VK (2015) Blumberg, CEACAM1 regulates TIM-3-mediated tolerance and exhaustion. Nature 517:386–390. https://doi.org/10.1038/nature13848
Kamizono S, Hanada T, Yasukawa H, Minoguchi S, Kato R, Minoguchi M, Hattori K, Hatakeyama S, Yada M, Morita S, Kitamura T, Kato H, Nakayama K, Yoshimura A (2001) The SOCS Box of SOCS-1 accelerates ubiquitin-dependent proteolysis of TEL-JAK2. J Biol Chem 276:12530–12538. https://doi.org/10.1074/jbc.M010074200
Kamran MZ, Patil P, GudeRP, (2013) Role of STAT3 in cancer metastasis and translational advances. Biomed Res Int. https://doi.org/10.1155/2013/421821
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH (2016) PubChem substance and compound databases. Nucleic Acids Res 44:D1202–D1213. https://doi.org/10.1093/nar/gkv951
Kortylewski M, Kujawski M, Wang T, Wei S, Zhang S, Pilon-Thomas S, Niu G, Kay H, Mulé J, Kerr WG, Jove R, Pardoll D, Yu H (2005) Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med 11:1314–1321. https://doi.org/10.1038/nm1325
Kortylewski M, Yu H (2008) Role of Stat3 in suppressing anti-tumor immunity. Curr Opin Immunol 20:228. https://doi.org/10.1016/J.COI.2008.03.010
Krasnov GS, Kudryavtseva AV, Snezhkina AV, Lakunina VA, Beniaminov AD, Melnikova NV, Dmitriev AA (2019) Pan-cancer analysis of TCGA data revealed promising reference genes for qPCR normalization. Front Genet. https://doi.org/10.3389/fgene.2019.00097
Krzyszczyk P, Acevedo A, Davidoff EJ, Timmins LM, Marrero-Berrios I, Patel M, White C, Lowe C, Sherba JJ, Hartmanshenn C, O’Neill KM, Balter ML, Fritz ZR, Androulakis IP, Schloss RS, Yarmush ML (2018) The growing role of precision and personalized medicine for cancer treatment. Technology 6:79–100. https://doi.org/10.1142/S2339547818300020
Larkin J, Ahmed CM, Wilson TD, Johnson HM (2013) Cells T. Front Immunol. https://doi.org/10.3389/fimmu.2013.00469
Liau NPD, Laktyushin A, Lucet IS, Murphy JM, Yao S, Whitlock E, Callaghan K, Nicola NA, Kershaw NJ, Babon JJ (2018) The molecular basis of JAK/STAT inhibition by SOCS1. Nat Commun 9:1558. https://doi.org/10.1038/s41467-018-04013-1
Liu Y, Hu X, Han C, Wang L, Zhang X, He X, Lu X (2015) Targeting tumor suppressor genes for cancer therapy. BioEssays 37:1277–1286. https://doi.org/10.1002/bies.201500093
Llanes-Fernández L, Álvarez-Goyanes RI, Arango-Prado MdelC, Alcocer-González JM, Mojarrieta JC, Pérez XE, López MO, Odio SF, Camacho-Rodríguez R, Guerra-Yi ME, Madrid-Marina V, Tamez-Guerra R (2006) Rodríguez-Padilla, Relationship between IL-10 and tumor markers in breast cancer patients. The Breast 15:482–489. https://doi.org/10.1016/j.breast.2005.09.012
Long L, Zhang X, Chen F, Pan Q, Phiphatwatchara P, Zeng Y, Chen H (2018) The promising immune checkpoint LAG-3: from tumor microenvironment to cancer immunotherapy. Genes Cancer 9:176–189. https://doi.org/10.18632/genesandcancer.180
Ludwig KF, Du W, Sorrelle NB, Wnuk-Lipinska K, Topalovski M, Toombs JE, Cruz VH, Yabuuchi S, Rajeshkumar NV, Maitra A, Lorens JB, Brekken RA (2018) Small-molecule inhibition of Axl targets tumor immune suppression and enhances chemotherapy in pancreatic cancer. Cancer Res 78:246–255. https://doi.org/10.1158/0008-5472.CAN-17-1973
Mercer F, Unutmaz D (2009) The biology of FoxP3: A key player in immune suppression during infections. In: Autoimmune diseases and cancer, pp 47–59. https://doi.org/10.1007/978-1-4419-1599-3_4
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. https://doi.org/10.1002/jcc.21256
Oft M (2014) IL-10 master switch from tumor-promoting inflammation to antitumor immunity. Cancer Immunol Res 2:194–199. https://doi.org/10.1158/2326-6066.CIR-13-0214
Osborn SL, Diehl G, Han S-J, Xue L, Kurd N, Hsieh K, Cado D, Robey EA, Winoto A (2010) Fas-associated death domain (FADD) is a negative regulator of T cell receptor-mediated necroptosis. Proc Natl Acad Sci 107:13034–13039. https://doi.org/10.1073/pnas.1005997107
Otto T, Sicinski P (2017) Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer 17:93–115. https://doi.org/10.1038/nrc.2016.138
Papoff G, Trivieri N, Crielesi R, Ruberti F, Marsilio S, Ruberti G (2010) FADD–calmodulin interaction: a novel player in cell cycle regulation. Biochim Biophys Acta Mol Cell Res 1803:898–911. https://doi.org/10.1016/j.bbamcr.2010.04.006
Paul MK, Mukhopadhyay AK (2004) Tyrosine kinase: role and significance in cancer. Int J Med Sci. https://doi.org/10.7150/ijms.1.101
Pawlus MR, Wang L, Hu C-J (2014) STAT3 and HIF1α cooperatively activate HIF1 target genes in MDA-MB-231 and RCC4 cells. Oncogene 33:1670–1679. https://doi.org/10.1038/onc.2013.115
Pestell KE (2003) Paul Workman on the challenges of cancer drug development. Drug Discov Today 8:775–777. https://doi.org/10.1016/S1359-6446(03)02838-1
Rankin E, Giaccia A (2016) The receptor tyrosine kinase AXL in cancer progression. Cancers (Basel) 8:103. https://doi.org/10.3390/cancers8110103
Sawyers C (2002) Rational therapeutic intervention in cancer: kinases as drug targets. Curr Opin Genet Dev 12:111–115. https://doi.org/10.1016/S0959-437X(01)00273-8
Schwartz DM, Kanno Y, Villarino A, Ward M, Gadina M, O’Shea JJ (2017) JAK inhibition as a therapeutic strategy for immune and inflammatory diseases. Nat Rev Drug Discov 17:78. https://doi.org/10.1038/NRD.2017.267
Schwartz DM, Kanno Y, Villarino A, Ward M, Gadina M, O’Shea JJ (2018) Erratum: JAK inhibition as a therapeutic strategy for immune and inflammatory diseases. Nat Rev Drug Discov 17:78–78. https://doi.org/10.1038/nrd.2017.267
Shao L, Hou W, Scharping NE, Vendetti FP, Srivastava R, Roy CN, Menk AV, Wang Y, Chauvin J-M, Karukonda P, Thorne SH, Hornung V, Zarour HM, Bakkenist CJ, Delgoffe GM (2019) Sarkar, IRF1 inhibits antitumor immunity through the upregulation of PD-L1 in the tumor cell. Cancer Immunol Res 7:1258–1266. https://doi.org/10.1158/2326-6066.CIR-18-0711
Sheikhpour E, Noorbakhsh P, Foroughi E, Farahnak S, Nasiri R, Neamatzadeh H (2018) A survey on the role of interleukin-10 in breast cancer: a narrative. Rep Biochem Mol Biol 7:30–37
Sriuranpong V, Park JI, Amornphimoltham P, Patel V, Nelkin BD, Gutkind JS (2003) Epidermal growth factor receptor-independent constitutive activation of STAT3 in head and neck squamous cell carcinoma is mediated by the autocrine/paracrine stimulation of the interleukin 6/gp130 cytokine system. Cancer Res 63:2948–2956. http://www.ncbi.nlm.nih.gov/pubmed/12782602
Stehelin D, Varmus HE, Bishop JM, Vogt PK (1976) DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA. Nature 260:170–173. https://doi.org/10.1038/260170a0
Sur I, Taipale J (2016) The role of enhancers in cancer. Nat Rev Cancer 16:483–493. https://doi.org/10.1038/nrc.2016.62
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C (2017) The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res 45:D362–D368. https://doi.org/10.1093/nar/gkw937
Tadesse S, Yu M, Kumarasiri M, Le BT, Wang S (2015) Targeting CDK6 in cancer: state of the art and new insights. Cell Cycle 14:3220–3230. https://doi.org/10.1080/15384101.2015.1084445
Tan KT, Yeh C-N, Chang Y-C, Cheng J-H, Fang W-L, Yeh Y-C, Wang Y-C, Hsu DS-S, Wu C-E, Lai J-I, Chang PM-H, Chen M-H, Lu M-L, Chen S-J, Chao Y, Hsiao M, Chen M-H (2020) PRKDC: new biomarker and drug target for checkpoint blockade immunotherapy. J Immunother Cancer 8:e000485. https://doi.org/10.1136/jitc-2019-000485
Tanegashima T, Togashi Y, Azuma K, Kawahara A, Ideguchi K, Sugiyama D, Kinoshita F, Akiba J, Kashiwagi E, Takeuchi A, Irie T, Tatsugami K, Hoshino T, Eto M, Nishikawa H (2019) Immune suppression by PD-L2 against spontaneous and treatment-related antitumor immunity. Clin Cancer Res 25:4808–4819. https://doi.org/10.1158/1078-0432.CCR-18-3991
Thomas SJ, Snowden JA, Zeidler MP, Danson SJ (2015) The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br J Cancer 113:365–371. https://doi.org/10.1038/bjc.2015.233
Tobelaim WS, Beaurivage C, Champagne A, Pomerleau V, Simoneau A, Chababi W, Yeganeh M, Thibault P, Klinck R, Carrier JC, Ferbeyre G, Ilangumaran S, Saucier C (2015) Tumour-promoting role of SOCS1 in colorectal cancer cells. Sci Rep 5:14301. https://doi.org/10.1038/srep14301
Torres N, Regge MV, Secchiari F, Friedrich AD, Spallanzani RG, Raffo Iraolagoitia XL, Núñez SY, Sierra JM, Ziblat A, Santilli MC, Gilio N, Almada E, Lauche C, Pardo R, Domaica CI, Fuertes MB, Madauss KP, Hance KW, Gloger IS, Zylberman V, Goldbaum FA, Zwirner NW (2020) Restoration of antitumor immunity through anti-MICA antibodies elicited with a chimeric protein. J Immunother Cancer 8:e000233. https://doi.org/10.1136/jitc-2019-000233
Twyman-Saint Victor C, Rech AJ, Maity A, Rengan R, Pauken KE, Stelekati E, Benci JL, Xu B, Dada H, Odorizzi PM, Herati RS, Mansfield KD, Patsch D, Amaravadi RK, Schuchter LM, Ishwaran H, Mick R, Pryma DA, Xu X, Feldman MD, Gangadhar TC, Hahn SM, Wherry EJ, Vonderheide RH, Minn AJ (2015) Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature 520:373–377. https://doi.org/10.1038/nature14292
Ullrich CI, Aloni R, Saeed MEM, Ullrich W, Efferth T (2019) Comparison between tumors in plants and human beings: mechanisms of tumor development and therapy with secondary plant metabolites. Phytomedicine 64:153081. https://doi.org/10.1016/j.phymed.2019.153081
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW (2013) Cancer genome landscapes. Science 339:1546–1558. https://doi.org/10.1126/science.1235122
Wang H, Brown J, Garcia CA, Tang Y, Benakanakere MR, Greenway T, Alard P, Kinane DF, Martin M (2011) The role of glycogen synthase kinase 3 in regulating IFN-β–mediated IL-10 production. J Immunol 186:675–684. https://doi.org/10.4049/jimmunol.1001473
Wehde BL, Rädler PD, Shrestha H, Johnson SJ, Triplett AA, Wagner K-U (2018) Janus kinase 1 plays a critical role in mammary cancer progression. Cell Rep 25:2192. https://doi.org/10.1016/J.CELREP.2018.10.063
Wei Y, Chen X, Yang J, Yao J, Yin N, Zhang Z, Li D, Zhu D, Zhou J (2019) DcR3 promotes proliferation and invasion of pancreatic cancer via a DcR3/STAT1/IRF1 feedback loop. Am J Cancer Res 9:2618–2633. http://www.ncbi.nlm.nih.gov/pubmed/31911850
Wuputra K, Ku C-C, Wu D-C, Lin Y-C, Saito S, Yokoyama KK (2020) Prevention of tumor risk associated with the reprogramming of human pluripotent stem cells. J Exp Clin Cancer Res 39:100. https://doi.org/10.1186/s13046-020-01584-0
Xia S, Ji R, Xu Y, Ni X, Dong Y, Zhan W (2017) Twisted gastrulation BMP signaling modulator 1 regulates papillary thyroid cancer cell motility and proliferation. J Cancer 8:2816–2827. https://doi.org/10.7150/jca.18482
Zhang Y, Zhaoyoung L (2017) STAT1 in cancer: friend or foe? Discov Med 24:19–29. https://www.discoverymedicine.com/Ying-Zhang/2017/08/stat1-in-cancer-friend-or-foe/
Yang S, Liu Y, Li M-Y, Ng CSH, Yang S, Wang S, Zou C, Dong Y, Du J, Long X, Liu L-Z, Wan IYP, Mok T, Underwood MJ, Chen GG (2017) FOXP3 promotes tumor growth and metastasis by activating Wnt/β-catenin signaling pathway and EMT in non-small cell lung cancer. Mol Cancer 16:124. https://doi.org/10.1186/s12943-017-0700-1
Yang L, Pang Y, Moses HL (2010) TGF-β and immune cells: an important regulatory axis in the tumor microenvironment and progression. Trends Immunol 31:220–227. https://doi.org/10.1016/j.it.2010.04.002
Yi T, Lee D-S, Jeon M-S, Kwon SW, Song SU (2012) Gene expression profile reveals that STAT2 is involved in the immunosuppressive function of human bone marrow-derived mesenchymal stem cells. Gene 497:131–139. https://doi.org/10.1016/j.gene.2012.01.073
Yu W, Hua Y, Qiu H, Hao J, Zou K, Li Z, Hu S, Guo P, Chen M, Sui S, Xiong Y, Li F, Lu J, Guo W, Luo G, Deng W (2020) PD-L1 promotes tumor growth and progression by activating WIP and β-catenin signaling pathways and predicts poor prognosis in lung cancer. Cell Death Dis 11:506. https://doi.org/10.1038/s41419-020-2701-z
Zhang J, Li H, Yu J-P, Wang SE, Ren X-B (2012) Role of SOCS1 in tumor progression and therapeutic application. Int J Cancer 130:1971–1980. https://doi.org/10.1002/ijc.27318
Zhang Y, Yang W, Wen G, Tang H, Wu C, Wu Y, Jing Z, Tang M, Liu G, Li D, Li Y, Deng Y (2019) High expression of PRKDC promotes breast cancer cell growth via p38 MAPK signaling and is associated with poor survival. Mol Genet Genomic Med. https://doi.org/10.1002/mgg3.908
Zhao Y, Yang W, Huang Y, Cui R, Li X, Li B (2018) Evolving roles for targeting CTLA-4 in cancer immunotherapy. Cell Physiol Biochem 47:721–734. https://doi.org/10.1159/000490025
Zhou X-M, Li W-Q, Wu Y-H, Han L, Cao X-G, Yang X-M, Wang H-F, Zhao W-S, Zhai W-J, Qi Y-M, Gao Y-F (2018) Intrinsic expression of immune checkpoint molecule TIGIT could help tumor growth in vivo by suppressing the function of NK and CD8+ T cells. Front Immunol. https://doi.org/10.3389/fimmu.2018.02821
Author Information
Department of Biotechnology, Delhi Technological University, New Delhi, India