Structural Characterization of complete Chloroplast Genome of Sporobolus helvolus (Poaceae)
Research Article | Published: 11 January, 2019
First Page: 35
Last Page: 38
Views: 3939
Keywords: Sporobolus helvolus, Chloroplast genome, Phylogeny, Poaceae
Abstract
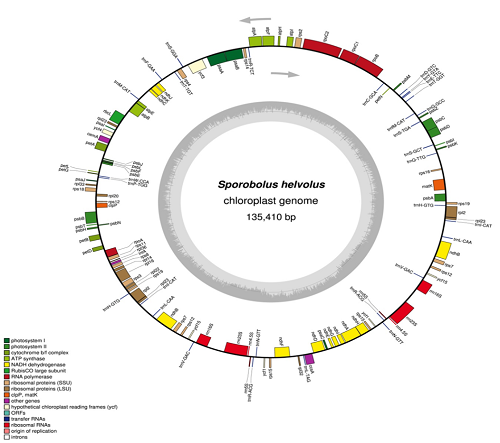
Sporobolus helvolus is perennial halophytic grass of family Poaceae. It is important grass for arid ecosystem because of its value as fodder and as controller of desertification. In present study complete chloroplast genome of S. helvolus was sequenced. The size of chloroplast genome is 135410 bp with overall GC content 38.4%. It exhibited regular quadripartite structure with 80719 bp of LSC region, 14699 bp of SSC region and 39992 bp of total IR region. A total of 127 genes were identified, including 89 coding genes, 30 tRNAs, 8 ribosomal RNAs and 1 pseudogene (ycf15). Phylogenetic analysis was performed using 14 other members representing major subfamilies of Poaceae and it clearly discriminate S. helvolus from other Sporobolus species.
References
- Andanje S and Ottichilo W (1999) Population status and feeding habits of the translocated sub‐population of Hunter's antelope or hirola (Beatragus hunteri, in Tsavo East National Park, Kenya. Afr J Ecol 37(1): 38-48.
- Bock R (2007) Structure, function, and inheritance of plastid genomes. Cell and molecular biology of plastids. Springer. Pp. 29-63
- Cai Z, Penaflor C, Kuehl JV, Leebens-Mack J, Carlson JE, Boore JL and Jansen RK (2006). Complete plastid genome sequences of Drimys, Liriodendron, and Piper: implications for the phylogenetic relationships of magnoliids. BMC Evol Biol 6(1): 77.
- Eckardt NA (2007). Chloroplast intron splicing mechanisms. Plant Cell 19(12): 3838.
- Goremykin VV, Hirsch-Ernst KI, Wölfl S and Hellwig FH (2003) Analysis of the Amborella trichopoda chloroplast genome sequence suggests that Amborella is not a basal angiosperm. Mol Biol Evol 20(9): 1499-1505.
- Gupta RK (1993) Utilization of salt tolerant plants from arid wastelands of Northwest India as fuel and fodder. Towards the rational use of high salinity tolerant plants: Vol. 2 Agriculture and forestry under marginal soil water conditions. Dordrecht: Springer Netherlands. H. Lieth and AA Al Masoom (Eds.) Pp. 247-257
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, and Guan X (2012) CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics 13(1): 715.
- Lowe TM and Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25(5): 955-964.
- Olejniczak SA, Łojewska E, Kowalczyk T and Sakowicz T (2016) Chloroplasts: state of research and practical applications of plastome sequencing. Planta 244(3): 517-527.
- Schmitz-Linneweber C, Maier RM, Alcaraz JP, Cottet A, Herrmann RG and Mache R. (2001). The plastid chromosome of spinach (Spinacia oleracea): complete nucleotide sequence and gene organization. Plant Mol Biol 45(3): 307-315.
- Shi C, Hu N, Huang H, Gao J, Zhao Y-J and Gao L-Z (2012) An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. Plos one 7(2): e31468.
- Steane DA (2005) Complete nucleotide sequence of the chloroplast genome from the Tasmanian blue gum, Eucalyptus globulus (Myrtaceae). DNA Res 12(3):215-220.
- Wyman SK, Jansen RK and Boore JL (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20(17): 3252-3255.
Author Information
Department of Biosciences, Saurashtra University, Rajkot 360 005, Gujarat, India