Withania somnifera phytochemicals possess SARS-CoV-2 RdRp and human TMPRSS2 protein binding potential
Prajapati Kumari Sunita, Singh Atul Kumar, Kushwaha Prem Prakash, Shuaib Mohd, Maurya Santosh Kumar, Gupta Sanjay, Senapati Sabyasachi, Singh Surya Pratap, Waseem Mohammad, Kumar Shashank
Research Articles | Published: 15 June, 2022
First Page: 701
Last Page: 720
Views: 3701
Keywords: SARS-CoV-2, Withania somnifera , Phytochemical, TMPRSS2, RNA dependent RNA polymerase
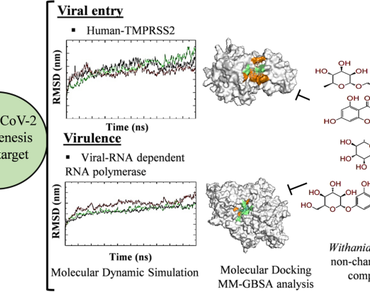
Abstract
References
Berger JP, SinhaRoy R, Pocai A, Kelly TM, Scapin G, Gao YD, Pryor KAD, Wu JK, Eiermann GJ, Xu SS, Zhang X, Tatosian DA, Weber AE, Thornberry NA, Carr RD (2017) A comparative study of the binding properties, dipeptidyl peptidase-4 (DPP-4) inhibitory activity and glucose-lowering efficacy of the DPP-4 inhibitors alogliptin, linagliptin, saxagliptin, sitagliptin and vildagliptin in mice. Endocrinol Diabetes Metab 1:1–8. https://doi.org/10.1002/edm2.2
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28(1):235–242. https://doi.org/10.1093/nar/28.1.235
Cai Z, Zhang G, Tang B, Liu Y, Fu X, Zhang X (2015) Promising Anti-influenza Properties of Active Constituent of Withania somnifera Ayurvedic Herb in Targeting Neuraminidase of H1N1 Influenza: Computational Study. Cell Biochem Biophys 72:727–739. https://doi.org/10.1007/s12013-015-0524-9
Casrouge A, Sauer AV, Barreira da Silva R, Tejera-Alhambra M, Sánchez-Ramón S, ICAReB, Cancrini C, Ingersoll MA, Aiuti A, Albert ML (2018) Lymphocytes are a major source of circulating soluble dipeptidyl peptidase 4. Clin Exp Immunol 194:166–179. https://doi.org/10.1111/cei.13163
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:1–13. https://doi.org/10.1038/srep42717
Dallakyan S (2008) PyRx-python prescription v. 0.8. The Scripps Research Institute. 2008; 2010
Das S, Sarmah S, Lyndem S, Singha Roy A (2021) An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J Biomol Struct Dyn 39:3347–3357. https://doi.org/10.1080/07391102.2020.1763201
Duke JA (1992) Database of phytochemical constituents of GRAS herbs and other economic plants. CRC Press, Boca Raton
El Khatabi K, Aanouz I, El-Mernissi R, Singh AK, Ajana MA, Lakhlifi T, Kumar S, Bouachrine M (2021) Integrated 3D-QSAR, molecular docking, and molecular dynamics simulation studies on 1,2,3-triazole based derivatives for designing new acetylcholinesterase inhibitors. Turk J Chem 45:647–660. https://doi.org/10.3906/kim-2010-34
Ezebuo FC, Kushwaha PP, Singh AK, Kumar S, Singh P (2019) In-silico methods of drug design: molecular simulations and free energy calculations. In: Kumar S, Egbuna C (eds) Phytochemistry: an in-silico and in-vitro update 2019. Springer, Singapore, pp 521–533
Ganguly B, Umapathi V, Rastogi SK (2018) Nitric oxide induced by Indian ginseng root extract inhibits Infectious Bursal Disease virus in chicken embryo fibroblasts in vitro. J Anim Sci Technol 60:1–5. https://doi.org/10.1186/s40781-017-0156-2
Gordon CJ, Tchesnokov EP, Feng JY, Porter DP, Götte M (2020) The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem 295:4773–4779. https://doi.org/10.1074/jbc.AC120.013056
Gupta S, Singh AK, Kushwaha PP, Prajapati KS, Shuaib M, Senapati S, Kumar S (2021a) Identification of potential natural inhibitors of SARS-CoV2 main protease by molecular docking and simulation studies. J Biomol Struct Dyn 39:4334–4345. https://doi.org/10.1080/07391102.2020.1776157
Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280. https://doi.org/10.1016/j.cell.2020.02.052
Iwata-Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, Nagata N (2019) TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol 93:1–15. https://doi.org/10.1128/JVI.01815-18
Jain J, Narayanan V, Chaturvedi S, Pai S, Sunil S (2018) In vivo evaluation of Withania somnifera-based Indian traditional formulation (Amukkara Choornam), against Chikungunya virus-induced morbidity and arthralgia. J Evid Based Integr Med 23:11–25. https://doi.org/10.1177/2156587218757661
Kempegowda PK, Zameer F, Narasimashetty CK, Kollur SP, Murari SK (2018) Inhibitory potency of Withania somnifera extracts against DPP-4: an in vitro evaluation. Afr J Tradit Complement Altern Med 15:11–25. https://doi.org/10.21010/ajtcam.v15i1.2
Kim SC, Schneeweiss S, Glynn RJ, Doherty M, Goldfine AB, Solomon DH (2015) Dipeptidyl peptidase-4 inhibitors in type 2 diabetes may reduce the risk of autoimmune diseases: a population-based cohort study. Ann Rheum Dis 74:1968–1975. https://doi.org/10.1136/annrheumdis-2014-205216
Kumar G, Patnaik R (2016) Exploring neuroprotective potential of Withania somnifera phytochemicals by inhibition of GluN2B-containing NMDA receptors: an in silico study. Med Hypotheses 92:35–43. https://doi.org/10.1016/j.mehy.2016.04.034
Kumar S, Kushwaha PP, Gupta S (2019) Emerging targets in cancer drug resistance. Cancer Drug Resist. https://doi.org/10.20517/cdr.2018.27. 2:161 – 77
Kushwaha PP, Vardhan PS, Kapewangolo P, Shuaib M, Prajapati SK, Singh AK, Kumar S (2019) Bulbine frutescens phytochemical inhibits notch signaling pathway and induces apoptosis in triple negative and luminal breast cancer cells. Life Sci 234:1–15. https://doi.org/10.1016/j.lfs.2019.116783
Kushwaha PP, Singh AK, Bansal T, Yadav A, Prajapati KS, Shuaib M, Kumar S (2020) Identification of natural inhibitors against SARS-CoV-2 Drugable targets using molecular docking, molecular dynamics Simulation, and MM-PBSA approach. Front Cell Infect Microbiol 11:1–17. https://doi.org/10.3389/fcimb.2021.730288
Kushwaha PP, Singh AK, Prajapati KS, Shuaib M, Fayez S, Bringmann G, Kumar S (2020a) Induction of apoptosis in breast cancer cells by naphthylisoquinoline alkaloids. Toxicol Appl Pharmacol 409:1–12. https://doi.org/10.1016/j.taap.2020.115297
Kushwaha PP, Singh AK, Shuaib M, Prajapati KS, Vardhan PS, Gupta S, Kumar S (2020b) 3-O-(E)-p-Coumaroyl betulinic acid possess anticancer activity and inhibit Notch signaling pathway in breast cancer cells and mammosphere. Chem Biol Interact 328:1–12. https://doi.org/10.1016/j.cbi.2020.109200
Kushwaha PP, Kumar R, Neog PR, Behara MR, Singh P, Kumar A, Prajapati KS, Singh AK, Shuaib M, Sharma AK, Pandey AK (2021b) Characterization of phytochemicals and validation of antioxidant and anticancer activity in some Indian polyherbal ayurvedic products. Vegetos 2021 34:286–299. https://doi.org/10.1007/s42535-021-00205-1
Kushwaha PP, Maurya SK, Singh A, Prajapati KS, Singh AK, Shuaib M, Kumar S (2021a) Bulbine frutescens phytochemicals as novel ABC-transporter inhibitor: a molecular docking and molecular dynamics simulation study. J Cancer Metastasis Treat 7:1–13. https://doi.org/10.20517/2394-4722.2020.92
Kushwaha PP, Singh AK, Prajapati KS, Shuaib M, Gupta S, Kumar S (2021) Phytochemicals present in Indian ginseng possess potential to inhibit SARS-CoV-2 virulence: a molecular docking and MD simulation study. Microb Pathog 157:1–11. https://doi.org/10.1016/j.micpath.2021.104954
Laskowski RA, Swindells MB (2011) LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 51:2778–2786. https://doi.org/10.1021/ci200227u
Li Y, But PP, Ooi VE (2005) Antiviral activity and mode of action of caffeoylquinic acids from Schefflera heptaphylla (L.) Frodin. Antiviral Res 68:1–9. https://doi.org/10.1016/j.antiviral.2005.06.004
Lu G, Hu Y, Wang Q, Qi J, Gao F, Li Y, Zhang Y, Zhang W, Yuan Y, Bao J, Zhang B, Shi Y, Yan J, Gao GF (2013) Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 500:227–231. https://doi.org/10.1038/nature12328
Maurya SK, Maurya AK, Mishra N, Siddique HR (2020) Virtual screening, ADME/T, and binding free energy analysis of anti-viral, anti-protease, and anti-infectious compounds against NSP10/NSP16 methyltransferase and main protease of SARS CoV-2. J Recept Signal Transduct Res 40:605–612. https://doi.org/10.1080/10799893.2020.1772298
Mundkinajeddu D, Sawant LP, Koshy R, Akunuri P, Singh VK, Mayachari A, Sharaf MH, Balasubramanian M, Agarwal A (2014) Development and validation of high performance liquid chromatography method for simultaneous estimation of flavonoid glycosides in Withania somnifera aerial parts. Int Sch Res Notices 2014:1–6. https://doi.org/10.1155/2014/351547
Nagarajan N, Yapp EKY, Le NQK, Yeh HY (2019) In silico screening of sugar alcohol compounds to inhibit viral matrix protein VP40 of Ebola virus. Mol Biol Rep 46:3315–3324. https://doi.org/10.1007/s11033-019-04792-w
Naveed M, Hejazi V, Abbas M, Kamboh AA, Khan GJ, Shumzaid M, Ahmad F, Babazadeh D, FangFang X, Modarresi-Ghazani F, WenHua L, XiaoHui Z (2018) Chlorogenic acid (CGA): a pharmacological review and call for further research. Biomed Pharmacother 97:67–74. https://doi.org/10.1016/j.biopha.2017.10.064
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open Babel: an open chemical toolbox. J Cheminform 3:1–14. https://doi.org/10.1186/1758-2946-3-33
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Schüttelkopf AW, van Aalten DM (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60:1355–1363. https://doi.org/10.1107/S0907444904011679
Schwarz S, Sauter D, Wang K, Zhang R, Sun B, Karioti A, Bilia AR, Efferth T, Schwarz W (2014) Kaempferol derivatives as antiviral drugs against the 3a channel protein of coronavirus. Planta Med 80:177–182. https://doi.org/10.1055/s-0033-1360277
Senapati S, Kumar S, Singh AK, Banerjee P, Bhagavatula S (2020) Assessment of risk conferred by coding and regulatory variations of TMPRSS2 and CD26 in susceptibility to SARS-CoV-2 infection in human. J Genet 99:1–5. https://doi.org/10.1007/s12041-020-01217-7
Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F (2020) Structural basis of receptor recognition by SARS-CoV-2. Nature 581:221–224. https://doi.org/10.1038/s41586-020-2179-y
Singh V, Singh B, Sharma A, Kaur K, Gupta AP, Salar RK, Hallan V, Pati PK (2017) Leaf spot disease adversely affects human health-promoting constituents and withanolide biosynthesis in Withania somnifera (L.) Dunal. J Appl Microbiol 122:153–165. https://doi.org/10.1111/jam.13314
Singh AK, Kushwaha PP, Prajapati KS, Shuaib M, Gupta S, Kumar S (2020) Identification of FDA approved drugs and nucleoside analogues as potential SARS-CoV-2 A1pp domain inhibitor: an in silico study. Comput Biol Med 130:1–10. https://doi.org/10.1016/j.compbiomed.2020.104185
Singh AK, Patel PK, Choudhary K, Joshi J, Yadav D, Jin JO (2020b) Quercetin and Coumarin Inhibit Dipeptidyl Peptidase-IV and Exhibits Antioxidant Properties: In Silico, In Vitro, Ex Vivo. Biomolecules 10:1–14. https://doi.org/10.3390/biom1002020b7
Singh AK, Shuaib M, Kushwaha PP, Prajapati KS, Sharma R, Kumar S (2021) In silico updates on lead identification for obesity and cancer. In: Kumar S, Gupta S (eds) Obesity and Cancer. Springer, Singapore, pp 257–277
Solerte SB, Di Sabatino A, Galli M, Fiorina P (2020) Dipeptidyl peptidase-4 (DPP4) inhibition in COVID-19. Acta Diabetol 57:779–783. https://doi.org/10.1007/s00592-020-01539-z
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc.21334
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ (2005) GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718. https://doi.org/10.1002/jcc.20291
Vankadari N, Wilce JA (2020) Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg Microbes Infect 9:601–604. https://doi.org/10.1080/22221751.2020.1739565
Verma AK, Maurya SK, Kumar A, Barik M, Yadav V, Umar B, Lawal M, Usman ZA, Adam MA, Awal B (2020) Inhibition of multidrug resistance property of Candida albicans by natural compounds of parthenium hysterophorus L. An in-silico approach. J Pharmacogn Phytochem 9:55–64. https://doi.org/10.22271/phyto.2020.v9.i3a.11480
Waseem M, Ahmad MK, Srivatava VK, Rastogi N, Serajuddin M, Kumar S, Mishra DP, Sankhwar SN, Mahdi AA (2017) Evaluation of miR-711 as novel biomarker in prostate cancer progression. Asian Pac J Cancer Prev 18:2185–2191. https://doi.org/10.22034/APJCP.2017.18.8.2185
Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, Wang Q, Xu Y, Li M, Li X, Zheng M, Chen L, Li H (2020) Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B 10:766–788. https://doi.org/10.1016/j.apsb.2020.02.008
Author Information
Molecular Signaling & Drug Discovery Laboratory, Department of Biochemistry, Central University of Punjab, Bathinda, India