A comparative analysis of deep learning-based techniques for miRNA prediction associated with mRNA sequences
*Article not assigned to an issue yet
Research Articles | Published: 13 April, 2024
Online ISSN : 2229-4473.
Website:www.vegetosindia.org
Pub Email: contact@vegetosindia.org
First Page: 0
Last Page: 0
Views: 1012
Keywords:
Convolutional neural network (CNN), Deep learning, Gene expression, Long short-term memory (LSTM) and miRNA sequences
Abstract
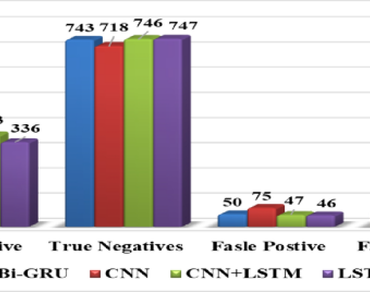
MicroRNAs (miRNAs) are short sequences of nucleotides, typically consisting of 21–25 base pairs, which play a crucial role in the regulation of genes throughout several biological processes. The identification of these miRNAs is challenging and intricate owing to their short read duration. Hence, the use of modern computational methodologies may provide significant benefits in accurately discerning these sequences. In recent years, there has been a growing use of computer methodologies for the categorization of diverse biological datasets. This work used publicly accessible miRNA sequences for the purpose of binary classification. Additionally, a dictionary was employed to numerically represent the nucleotide sequences, which were of a consistent length of 22 nucleotides. Various deep learning approaches, including Bidirectional Gated Recurrent Unit (Bi-GRU), Convolutional Neural Network (CNN), a mix of CNN and Long Short-Term Memory (LSTM), and LSTM, were used in the research investigation. All of the models exhibited much higher efficiency in comparison to the models documented in existing literature. Additionally, it was noted that the hybrid model combining Convolutional Neural Network (CNN) and Long Short-Term Memory (LSTM) has superior performance compared to the other models, with the maximum classification accuracy of 92.8% on the testing dataset. This hybrid model presented in this study represents the first development of a classification model specifically designed for the categorization of miRNA sequences derived from either plant or animal sources. Our developed hybrid model efficiently classify the data as it uses two different algorithms in model building.
(*Only SPR Members can get full access. Click Here to Apply and get access)
References
Ahmed B, Rai A, Iquebal MA, Jaiswal S (2021) Comparative analysis of machine learning and deep learning-based classification for abiotic stress proteins. Ind J Agric Sci 91(6):861–866
Ahmed B, Haque A, Iquebal MA, Jaiswal S, Angadi UB, Kumar D, Rai A (2023) Deepaprot: deep learning based abiotic stress protein sequence classification and identification tool in cereals. Front Plant Sci. https://doi.org/10.3389/fpls.2022.1008756
Barman M, Samanta S, Ahmed B, Dey S, Chakraborty S, Deeksha MG, Dutta S, Samanta A, Tarafdar J, Roy D (2023) Transcription dynamics of heat-shock proteins (Hsps) and endosymbiont titres in response to thermal stress in whitefly, Bemisia tabaci (Asia-I). Front Physiol 13:2762
Cai Y, Wang J, Deng L (2020) SDN2GO : an integrated deep learning model for protein function prediction. Front Bioeng Biotecnol 8:1–11
Das B, Torman S (2020) Classifying protein sequences using convolutional neural network. Bitlis Eren Üniversitesi Fen Bilimleri Dergisi 9(4):1663–1671
Gilani N, ArabiBelaghi R, Aftabi Y, Faramarzi E, Edgünlü T, Somi MH (2022) Identifying potential mirna biomarkers for gastric cancer diagnosis using machine learning variable selection approach. Front Genet 12:1–10
Haque MA, Marwaha S, Arora A, Paul RK, Hooda KS, Sharma A, Grover M (2021) Image-based identification of maydis leaf blight disease of maize (Zea mays) using deep learning. Ind J Agric Sci 91(9):1362–1367
Haque MA, Marwaha S, Deb CK, Nigam N, Arora A, Hooda KS, Soujanya PL, Aggarwal SS, Lall B, Kumar M, Islam S, Panwar M, Kumar P, Agarwal RC (2022) Deep learning-based approach for identification of diseases of maize crop. Sci Rep 12(6334):1–14
He L, Hannon GJ (2004) MicroRNAs: small rnas with a big role in gene regulation. Nat Rev Genet 5(7):522–531
Helwak A, Tollervey D (2014) Mapping the human miRNA interactome by clash reveals frequent noncanonical binding. Cell 153:654–665
Jayasundara S, Lokuge S, Ihalagedara P, and Herath D. (2021). Machine learning for plant microRNA prediction: A systematic review. arXiv:2106.15159. pp 1–15
Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42:D68–D73
Lee D, Lim M, Park H, Kang Y, Park JS, Jang GJ, Kim JH (2017) Long short-term memory recurrent neural network-based acoustic model using connectionist temporal classification on a large-scale training corpus. China Commun 14(9):23–31
Li T, Hua M, Wu X (2020) A hybrid cnn-lstm model for forecasting particulate matter (PM2.5). IEEE Access 8:26933–26940
Mendell JE, Olson EN (2012) MicroRNAs in stress signaling and human disease. NIH Public Access. Bone 148(6):1172–1187. https://doi.org/10.1016/j.cell.2012.02.005
Menor M, Ching T, Zhu X, Garmire D, Garmire LX (2014) Midmark: a site-level and UTR-level classifier for miRNA target prediction. Genome Biol 15:500
Min S, Lee B, Yoon S (2022) Targetnet: functional microRNA target prediction with deep neural networks. Bioinformatics 38(3):671–677
Patil A, Rane M (2021) Convolutional neural networks: an overview and its applications in pattern recognition. Smart Innov, Syst Technol 195:21–30
Pla A, Zhong X, Rayner S (2018) miRAW: A deep learning-based approach to predict microRNA targets by analyzing whole microRNA transcripts. In PLoS Comput Biol 14(7):1–32
Acknowledgements
The authors are grateful to ICAR-IASRI, New Delhi, Galgotias University, Greater Noida, India and 4University of Nebraska–Lincoln, USA for providing all the required facilities.
Author Information
ICAR-Indian Agricultural Statistics Research Institute, New Delhi, India
ahmedbulbul52@gmail.com