Identification and co-expression analysis of temperature responsive circRNAs in tea plant
*Article not assigned to an issue yet
Research Articles | Published: 16 April, 2024
Online ISSN : 2229-4473.
Website:www.vegetosindia.org
Pub Email: contact@vegetosindia.org
First Page: 0
Last Page: 0
Views: 998
Keywords:
Circular RNAs, Non-coding RNAs, Weighted gene co-expression network analysis, Competing endogenous RNA (ceRNA), Endogenous target mimics, Transcription factors
Abstract
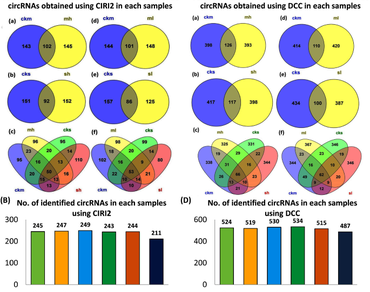
Circular RNAs (circRNAs) are recently known to regulate various biological processes associated with developmental transitions and stress condition in plants. In this study, a total of 763 circRNAs were identified in tea plants during temperature stress conditions, among which 60 circRNAs are found to be differentially expressed (DE). Gene set enrichment analysis (GSEA) of Gene Ontology (GO) revealed that parental genes of DE circRNAs were activated in response to heat, response to temperature stimulus, protein binding, response to stress, etc. Weighted gene co-expression network analysis (WGCNA) showed that 7 circRNAs co-expressed with 601 genes that are positively correlated in high temperature stresses, whereas 4 circRNAs co-expressed with 169 genes that showed positive correlation to low temperature stresses. A total of 42 DE circRNAs were predicted to act as competing endogenous RNA (ceRNA), sponging the miRNA activity by acting as endogenous target mimics (eTMs) for different transcription factors (TFs) like HSF, MYB, WRKY, etc. This study identified temperature responsive circRNAs in tea plants and also establishes a foundation for further research on candidate circRNAs in temperature stress conditions.
(*Only SPR Members can get full access. Click Here to Apply and get access)
References
Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281(49):37636–37645. https://doi.org/10.1074/jbc.M605895200
Babbar R, Tiwari LD, Mishra RC, Shimphrui R, Singh AA, Goyal I et al (2023) Arabidopsis plants overexpressing additional copies of heat shock protein Hsp101 showed high heat tolerance and endo-gene silencing. Plant Sci 330:111639. https://doi.org/10.1016/j.plantsci.2023.111639
Barrett SP, Salzman J (2016) Circular RNAs: analysis, expression and potential functions. Development 143(11):1838–1847. https://doi.org/10.1242/dev.128074
Baruah PM, Kashyap P, Krishnatreya DB, Bordoloi KS, Gill SS, Agarwala N (2021b) Identification and functional analysis of drought responsive lncRNAs in tea plant. Plant Gene 27:100311. https://doi.org/10.1016/j.plgene.2021.100311
Baruah PM, Krishnatreya DB, Bordoloi KS, Gill SS, Agarwala N (2021a) Genome wide identification and characterization of abiotic stress responsive lncRNAs in Capsicum annuum. Plant Physiol Biochem 162:221–236. https://doi.org/10.1016/j.plaphy.2021.02.031
Baruah PM, Bordoloi KS, Gill SS, Agarwala N (2023) CircRNAs responsive to winter dormancy and spring flushing conditions of tea leaf buds. Plant Sci 336:111828. https://doi.org/10.1016/j.plantsci.2023.111828
Boehm R, Cash SB, Anderson BT, Ahmed S, Griffin TS, Robbat A Jr et al (2016) Association between empirically estimated monsoon dynamics and other weather factors and historical tea yields in China: results from a yield response model. Climate 4(2):20. https://doi.org/10.3390/cli4020020
Bordoloi KS, Baruah PM, Agarwala N (2023) Identification of circular RNAs in tea plant during Helopeltis Theivora infestation. Plant Stress 8:100150. https://doi.org/10.1016/j.stress.2023.100150
Bordoloi KS, Baruah PM, Tanti B, Gill SS, Agarwala N (2022) Helopeltis Theivora Responsive Transcriptomic Reprogramming uncovers long non-coding RNAs as possible regulators of primary and secondary metabolism in Tea Plant. J Plant Growth Regul 1–26. https://doi.org/10.1007/s00344-022-10893-x
Chaudhry S, Sidhu GPS (2022) Climate change regulated abiotic stress mechanisms in plants: a comprehensive review. Plant Cell Rep 41:1–31. https://doi.org/10.1007/s00299-021-02759-5
Chen LL (2016) The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 17(4):205–211. https://doi.org/10.1038/nrm.2015.32
Chen L, Song Y, Li S, Zhang L, Zou C, Yu D (2012) The role of WRKY transcription factors in plant abiotic stresses. Biochimica et Biophysica Acta (BBA)-Gene Regulatory mechanisms. 1819(2):120–128. https://doi.org/10.1016/j.bbagrm.2011.09.002
Chen L, Zhang P, Fan Y, Lu Q, Li Q, Yan J, Muehlbauer GJ, Schnable PS, Dai M, Li L (2018) Circular RNAs mediated by transposons are associated with transcriptomic and phenotypic variation in maize. New Phytol 217(3):1292–1306. https://doi.org/10.1111/nph.14901
Chen Y, Chen Z, Kang J, Kang D, Gu H, Qin G (2013) AtMYB14 regulates cold tolerance in Arabidopsis. Plant Mol Biology Report 31:87–97. https://doi.org/10.1007/s11105-012-0481-z
Cheng J, Metge F, Dieterich C (2016) Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics 32(7):1094–1096. https://doi.org/10.1093/bioinformatics/btv656
Chu Q, Bai P, Zhu X, Zhang X, Mao L, Zhu QH et al (2020) Characteristics of plant circular RNAs. Brief Bioinform 21(1):135–143. https://doi.org/10.1093/bib/bby111
Chu Q, Ding Y, Xu X, Ye CY, Zhu QH, Guo L, Fan L (2022) Recent origination of circular RNAs in plants. New Phytol 233(1):515–525. https://doi.org/10.1111/nph.17798
Chu Q, Zhang X, Zhu X, Liu C, Mao L, Ye C, Zhu QH et al (2017) PlantcircBase: a database for plant circular RNAs. Mol Plant 10:1126–1128. https://doi.org/10.1016/j.xplc.2022.100343
Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic acids research, 39(web server issue). W155–W159. https://doi.org/10.1093/nar/gkr319
Darbani B, Noeparvar S, Borg S (2016) Identification of circular RNAs from the parental genes involved in multiple aspects of Cellular Metabolism in Barley. Front Plant Sci 7:776. https://doi.org/10.3389/fpls.2016.00776
Ding Y, Yang S (2022) Surviving and thriving: how plants perceive and respond to temperature stress. Dev Cell 57(8):947–958. https://doi.org/10.1016/j.devcel.2022.03.010
Eisenhardt B (2013) Small heat shock proteins: recent developments. Biomol Concepts 4(6):583–595. https://doi.org/10.1515/bmc-2013-0028
Fatima B, Batcho AA, Sandhu ZY, Sarwar MB, Hassan S, Rashid B (2022) Heat shock proteins (HSP70) gene: Plant Transcriptomic Oven in the Hot Desert. Plant Response Mech Abiotic Stresses. https://doi.org/10.5772/intechopen.105391
Gao Y, Wang J, Zhao F (2015) Genome Biol 16(1):1–16. https://doi.org/10.1186/s13059-014-0571-3. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification
Gao Y, Zhang J, Zhao F (2018) Circular RNA identification based on multiple seed matching. Brief Bioinform 19(5):803–810. https://doi.org/10.1093/bib/bbx014
Gao Z, Li J, Luo M, Li H, Chen Q et al (2019) Characterization and cloning of grape circular RNAs identified the cold resistance-related Vv-circATS1. Plant Physiol 180(2):966–985. https://doi.org/10.1104/pp.18.01331
Giesguth M, Sahm A, Simon S, Dietz KJ (2015) Redox-dependent translocation of the heat shock transcription factor AtHSFA8 from the cytosol to the nucleus in Arabidopsis thaliana. FEBS Lett 589:718–725. https://doi.org/10.1016/j.febslet.2015.01.039
Guo M, Liu JH, Ma X, Luo DX, Gong ZH, Lu MH (2016) The Plant Heat Stress Transcription Factors (HSFs): structure, regulation, and function in response to Abiotic stresses. Front Plant Sci 7:114. https://doi.org/10.3389/fpls.2016.00114
Guria A, Sharma P, Srikakulam N, Baby A, Natesan S, Pandi G (2022) Cost-effective transcriptome-wide profiling of circular RNAs by the Improved-tdMDA-NGS method. Front Mol Biosci 9:886366. https://doi.org/10.3389/fmolb.2022.886366
Guria A, Sharma P, Natesan S, Pandi G (2020) Circular RNAs—the road less traveled. Front Mol Biosci 6:146. https://doi.org/10.3389/fmolb.2019.00146
Guria, A., Velayudha Vimala Kumar, K., Srikakulam, N., Krishnamma, A., Chanda, S.,Sharma, S.,… Pandi, G. (2019). Circular RNA profiling by Illumina sequencing via template-dependent multiple displacement amplification. BioMed Research International,2019. https://doi.org/10.1155/2019/2756516
Han D, Yang F, Yang ZQ, Jin ZF (2016) Effects of high temperature stress and recovery on photosynthesis and stress tolerance of tea leaves. Chin J Agrometeorology 37:297:306
Huang J, Wang Y, Yu J, Li F, Yi L, Li Y, Xie N, Wu Q, Samarina L, Tong W, Xia E (2023) Evolutionary Landscape of Tea Circular RNAs and its contribution to Chilling Tolerance of Tea Plant. Int J Mol Sci 24(2):1478. https://doi.org/10.3390/ijms24021478
Huang X, Zhang H, Guo R, Wang Q, Liu X, Kuang W, Song H, Liao J, Huang Y, Wang Z (2021) Systematic identification and characterization of circular RNAs involved in flag leaf senescence of rice. Planta 253(2):26. https://doi.org/10.1007/s00425-020-03544-6
Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32(5):453–461. https://doi.org/10.1038/nbt.2890
Jeyaraj A, Liu S, Zhang X, Zhang R, Shangguan M, Wei C (2017b) Genome-wide identification of microRNAs responsive to Ectropis oblique feeding in tea plant (Camellia sinensis L). Sci Rep 7(1):13634. https://doi.org/10.1038/s41598-017-13692-7
Jeyaraj A, Zhang X, Hou Y, Shangguan M, Gajjeraman P, Li Y, Wei C (2017a) Genome-wide identification of conserved and novel microRNAs in one bud and two tender leaves of tea plant (Camellia sinensis) by small RNA sequencing, microarray-based hybridization and genome survey scaffold sequences. BMC Plant Biol 17(1):212. https://doi.org/10.1186/s12870-017-1169-1
Jiang S, Yang Y, Guo D, Wang Y, Huang H, Yang et al (2022) Functional annotation of circRNAs of tea leaves during infection by the tea leaf spot pathogen Didymella Segeticola. PhytoFrontiers™ 2(1):80–83. https://doi.org/10.1094/PHYTOFR-08-21-0054-A
Kalwan G, Gill SS, Priyadarshini P, Gill R, Yadava YK, Yadav S et al (2023) Approaches for identification and analysis of plant circular RNAs and their role in stress responses. Environ Exp Bot 205:105099. https://doi.org/10.1016/j.envexpbot.2022.105099
Krishnatreya DB, Agarwala N, Gill SS, Bandyopadhyay T (2021) Understanding the role of miRNAs for improvement of tea quality and stress tolerance. J Biotechnol 328:34–46. https://doi.org/10.1016/j.jbiotec.2020.12.019
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9:559. https://doi.org/10.1186/1471-2105-9-559
Li B, Gao K, Ren H, Tang W (2018) Molecular mechanisms governing plant responses to high temperatures. J Integr Plant Biol 60(9):757–779. https://doi.org/10.1111/jipb.12701
Li B, Feng C, Zhang W, Sun S, Yue D, Zhang X, Yang X (2023) Comprehensive non-coding RNA analysis reveals specific lncRNA/circRNA–miRNA–mRNA regulatory networks in the cotton response to drought stress. Int J Biol Macromol 253:126558. https://doi.org/10.1016/j.ijbiomac.2023.126558
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinf (Oxford England) 30(7):923–930. https://doi.org/10.1093/bioinformatics/btt656
Liu R, Ma Y, Guo T, Li G (2023) Identification, biogenesis, function, and mechanism of action of circular RNAs in plants. Plant Commun 4(1):100430. https://doi.org/10.1016/j.xplc.2022.100430
Liu SC, Xu YX, Ma JQ, Wang WW, Chen W, Huang DJ, Fang J, Li XJ, Chen L (2016) Small RNA and degradome profiling reveals important roles for microRNAs and their targets in tea plant response to drought stress. Physiol Plant 158(4):435–451. https://doi.org/10.1111/ppl.12477
Liu S, Mi X, Zhang R, An Y, Zhou Q, Yang T, Xia X, Guo R, Wang X, Wei C (2019) Integrated analysis of miRNAs and their targets reveals that miR319c/TCP2 regulates apical bud burst in tea plant (Camellia sinensis). Planta 250(4):1111–1129. https://doi.org/10.1007/s00425-019-03207-1
Liu ZQ, Shi LP, Yang S, Qiu SS, Ma XL, Cai JS et al (2021) A conserved double-W box in the promoter of CaWRKY40 mediates autoregulation during response to pathogen attack and heat stress in pepper. Mol Plant Pathol 22(1):3–18. https://doi.org/10.1111/mpp.13004
Love MI, Huber W, Anders S (2014) Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550. https://doi.org/10.1186/s13059-014-0550-8
Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H, Zhao Q, Zhou C, Zhao Y, Lu D, Luo J, Wang Y, Tian Q, Feng Q, Huang T, Han B (2015) Transcriptome-wide investigation of circular RNAs in rice. RNA (New York, N.Y.), 21(12), 2076–2087. https://doi.org/10.1261/rna.052282.115
McLoughlin F, Basha E, Fowler ME, Kim M, Bordowitz J, Katiyar-Agarwal S et al (2016) Class I and II small heat shock proteins together with HSP101 protect protein translation factors during heat stress. Plant Physiol. 172, 1221–1236. https://doi.org/10.1104/pp.16.00536
Misir S, Wu N, Yang BB (2022) Specific expression and functions of circular RNAs. Cell Death Differ 29(3):481–491. https://doi.org/10.1038/s41418-022-00948-7
Pan T, Sun X, Liu Y, Li H, Deng G, Lin H, Wang S (2018) Heat stress alters genome-wide profiles of circular RNAs in Arabidopsis. Plant Mol Biol 96:217–229. https://doi.org/10.1007/s11103-017-0684-7
Pasquali G, Biricolti S, Locatelli F, Baldoni E, Mattana M (2008) Osmyb4 expression improves adaptive responses to drought and cold stress in transgenic apples. Plant Cell Rep 27(10):1677–1686. https://doi.org/10.1007/s00299-008-0587-9
Qu H, Liu Y, Jiang H, Liu Y, Song W, Chen L (2021) Identification and characterization of miRNAs associated with sterile flower buds in the tea plant based on small RNA sequencing. Hereditas 158(1):26. https://doi.org/10.1186/s41065-021-00188-8
Raza A, Razzaq A, Mehmood SS, Zou X, Zhang X, Lv Y, Xu J (2019) Impact of climate change on crops adaptation and strategies to tackle its outcome: a review. Plants 8(2):34. https://doi.org/10.3390/plants8020034
Salzman J (2016) Circular RNA expression: its potential regulation and function. Trends Genet 32(5):309–316. https://doi.org/10.1016/j.tig.2016.03.002
Seth R, Maritim TK, Parmar R, Sharma RK (2021) Underpinning the molecular programming attributing heat stress associated thermotolerance in tea (Camellia sinensis (L.) O. Kuntze). Hortic Res 8. https://doi.org/10.1038/s41438-021-00532-z
Shang Y, Yan L, Liu ZQ, Cao Z, Mei C, Xin Q wu, Du FQ, Jiang SY, Zhang T, Zhao XF, Sun R, Liu HL, Yu R, Zhang YT (2010) DP The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell, 22, 1909–1935. https://doi.org/10.1105/tpc.110.073874
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Shen J, Zhang D, Zhou L, Zhang X, Liao J, Duan Y, Wen B, Ma Y, Wang Y, Fang W, Zhu X (2019) Transcriptomic and metabolomic profiling of Camellia sinensis L. Cv. ‘Suchazao’ exposed to temperature stresses reveals modification in protein synthesis and photosynthetic and anthocyanin biosynthetic pathways. Tree Physiol 39(9):1583–1599. https://doi.org/10.1093/treephys/tpz059
Sun P, Cheng C, Lin Y, Zhu Q, Lin J, Lai Z (2017) Combined small RNA and degradome sequencing reveals complex microRNA regulation of catechin biosynthesis in tea (Camellia sinensis). PLoS ONE 12(2):e0171173. https://doi.org/10.1371/journal.pone.0171173
Tan J, Zhou Z, Niu Y, Sun X, Deng Z (2017) Identification and functional characterization of tomato circRNAs derived from genes involved in fruit pigment accumulation. Sci Rep 7(1):8594. https://doi.org/10.1038/s41598-017-08806-0
Tong W, Yu J, Hou Y, Li F, Zhou Q, Wei C, Bennetzen JL (2018) Circular RNA architecture and differentiation during leaf bud to young leaf development in tea (Camellia sinensis). Planta 248(6):1417–1429. https://doi.org/10.1007/s00425-018-2983-x
Wang H, Zhong L, Fu X, Huang S, Zhao D, He H, Chen X (2023) Physiological analysis reveals the mechanism of accelerated growth recovery for rice seedlings by nitrogen application after low temperature stress. Front Plant Sci 14:1133592. https://doi.org/10.3389/fpls.2023.1133592
Wang J, Lin J, Wang H, Li X, Yang Q, Li H, Chang Y (2018) Identification and characterization of circRNAs in Pyrus Betulifolia Bunge under drought stress. PLoS ONE 13(7):e0200692. https://doi.org/10.1371/journal.pone.0200692
Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J (2014) Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE 9(3):e90859. https://doi.org/10.1371/journal.pone.0095116
Wang W, Wang J, Wei Q, Li B, Zhong X, Hu et al (2019) Transcriptome-wide identification and characterization of circular RNAs in leaves of Chinese cabbage (Brassica rapa L. ssp. pekinensis) in response to calcium deficiency-induced tip-burn. Sci Rep 9(1):1–9. https://doi.org/10.1038/s41598-019-51190-0
Wang Y, Yang M, Wei S, Qin F, Zhao H, Suo B (2017) Identification of circular RNAs and their targets in leaves of Triticum aestivum L. under dehydration stress. Front Plant Sci 7:2024. https://doi.org/10.3389/fpls.2016.02024
Wu HJ, Ma YK, Chen T, Wang M, Wang XJ (2012) PsRobot: a web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res W22–W28 40(Web Server issue). https://doi.org/10.1093/nar/gks554
Xia, E. H., Li, F. D., Tong, W., Li, P. H., Wu, Q., Zhao, H. J.,… Wan, X. C. (2019).Tea Plant Information Archive: a comprehensive genomics and bioinformatics platform for tea plant. Plant biotechnology journal, 17(10), 1938–1953. https://doi.org/10.1111/pbi.13111
Yamada K, Fukao Y, Hayashi M, Fukazawa M, Suzuki I, Nishimura M (2007) Cytosolic HSP90 regulates the heat shock response that is responsible for heat acclimation in Arabidopsis thaliana. J Biol Chem 282(52):37794–37804. https://doi.org/10.1074/jbc.M707168200
Yang Y, Jiang X, Shi J, Wang Y, Huang H, Yang Y et al (2022) Functional annotation of circRNAs in tea leaves after infection by the tea leaf spot pathogen. Lasiodiplodia Theobromae Phytopathology® 112(2):460–463. https://doi.org/10.1094/PHYTO-05-21-0184-A
Ye CY, Chen L, Liu C, Zhu QH, Fan L (2015) Widespread noncoding circular RNA s in plants. New Phytol 208(1):88–95. https://doi.org/10.1111/nph.13585
Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16(5):284–287. https://doi.org/10.1089/omi.2011.0118
Zhang P, Fan Y, Sun X, Chen L, Terzaghi W, Bucher et al (2019) A large-scale circular RNA profiling reveals universal molecular mechanisms responsive to drought stress in maize and Arabidopsis. Plant J 98(4):697–713. https://doi.org/10.1111/tpj.14267
Zhang Y, Zhu X, Chen X, Song C, Zou Z, Wang et al (2014) Identification and characterization of cold-responsive microRNAs in tea plant (Camellia sinensis) and their targets using high-throughput sequencing and degradome analysis. BMC Plant Biol 14(1):1–18. https://doi.org/10.1186/s12870-014-0271-x
Zhao W, Cheng Y, Zhang C, You Q, Shen X, Guo W, Jiao Y (2017) Genome-wide identification and characterization of circular RNAs by high throughput sequencing in soybean. Sci Rep 7(1):5636. https://doi.org/10.1038/s41598-017-05922-9
Zhou R, Zhu Y, Zhao J, Fang Z, Wang S, Yin J, Chu Z, Ma D (2017) Transcriptome-wide identification and characterization of Potato circular RNAs in response to Pectobacterium carotovorum subspecies brasiliense infection. Int J Mol Sci 19(1):71. https://doi.org/10.3390/ijms19010071
Zhou Y, Xu F, Shao Y, He J (2022) Regulatory Mechanisms of Heat Stress Response and thermomorphogenesis in plants. Plants 11(24):3410. https://doi.org/10.3390/plants11243410
Zhu YX, Jia JH, Yang L et al (2019) Identification of cucumber circular RNAs responsive to salt stress. BMC Plant Biol 19:164. https://doi.org/10.1186/s12870-019-1712-3
Zuo J, Wang Q, Zhu B, Luo Y, Gao L (2016) Deciphering the roles of circRNAs on chilling injury in tomato. Biochem Biophys Res Commun 479(2):132–138. https://doi.org/10.1016/j.bbrc.2016.07.032
Acknowledgements
We are thankful to DST, Govt. of India for providing DST-FIST support to the Department of Botany and DST-PURSE support to Gauhati University, where this research work was carried out. KSB thankfully acknowledge Council of Scientific and Industrial Research (CSIR) for CSIR-JRF fellowship under award letter number09/059(0064)-2018-EMR-I.
Author Information