Evaluation of genetic diversity in rice (Oryza sativa L. ssp. Indica) accessions using SSR marker
Research Articles | Published: 09 April, 2022
First Page: 961
Last Page: 968
Views: 3443
Keywords: SSR, PIC, Structure, AMOVA
Abstract
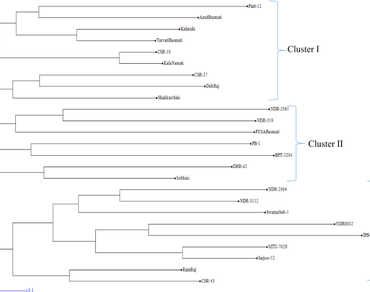
Genetic relationship among 25 rice accessions have been evaluated using 21 SSR markers. A total of 87 alleles were produced by 21 polymorphic SSRs with an average of 4 alleles per SSRs. The SSR RM514 produced maximum number of alleles (9) followed by RM125 (7) and RM271 and RM447 (6). The PIC value varied from 0.07 (RM507) to 0.83 (RM514) with an average of 0.46. Jaccard’s dissimilarity coefficient ranged 0.10–0.89 with an average of 0.65. The highest genetic dissimilarity was observed between Dub Raj & IPB—1 (0.89), and Pant—12 & IPB—1 (0.89) and minimum dissimilarity observed between Kalanamak and CSR—10 (0.10). By evaluating data from clustering, dissimilarity and admixture nature the most diverse accessions were IPB—1, Pant—12, Dub Raj, Ram Raj, NDR 2064 and Sobhini. The accessions IPB—1, NDR—8002, BPT—5204, NDR—2065, PB—1, PUSA Basmati, and NDR—359 was found highly diverse as these accessions showed highest dissimilarity with respect to rest of the accessions. The NJ clustering pattern revealed 3 clusters and supported by 3 subpopulations determined from STRUCTURE analysis. The analysis of molecular variance (AMOVA) showed that 85% of allelic diversity was found within the population.
References
Ahmed S, Anik TR, Islam A, Uddin I and Haque MS (2019) Screening of Some Rice (Oryza sativa L.) Genotypes for Salinity Tolerance using Morphological and Molecular Markers. Biosciences Biotechnology Research Asia 16(2):377–390
Borba TCO, Brondani RPV, Rangel PHN, Brondani C (2009) Microsatellite marker-mediated analysis of the EMBRAPA rice core collection genetic diversity. Genetica 137(3):293–304
Chakravarthi, BK, Naravaneni R (2006) SSR marker based DNA fingerprinting and diversity study in rice (Oryza sativa. L). African Journal of Biotechnology, 5(9)
Earl DA, vonHoldt BM (2012) Structure Harvester: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
Embate MVG, Calayugan MIC, Gentallan RP, Cruz PCS, Hernandez JE and Borromeo TH (2021) Genetic diversity of selected pigmented traditional rice (Oryza sativa L.) varieties from Mindanao, Philippines using agromorphological traits and simple sequence repeats markers. J Crop Sci Biotechnol 24(3):259–277
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol 14(8):2611–2620
Excoffier L, Hofer T, Foll M (2009) Detecting loci under selection in a hierarchically structured population. Heredity 103:285–298
Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S (2005) Genetic structure and diversity in Oryza sativa L. Genetics 169(3):1631–1638
Hoshino A A, Bravo J P, Nobile P M, Morelli K A (2012). Microsatellites as tools for genetic diversity analysis, genetic diversity in microorganisms. InTech Open: 382
Hossain MZ, Rasul MG, Ali MS, Iftekharuddaula KM, Mian MAK (2007) Molecular characterization and genetic diversity in fine grain and aromatic landraces of rice using microsatellite markers. Bangladesh J Genet Plant Breeding 20(2):1–10
Huang M, Xie F, Chen L, Zhao X, Jojee L, Madonna M (2010) Comparative analysis of genetic diversity and structure in rice using ILP and SSR markers. Rice Sci 17(4):257–268
Islam MZ, Khalequzzaman M, Prince MFRK, Siddique MA, Rashid ESMH, Ahmed MSU, Pittendrigh, BR and Ali MP (2018). Diversity and population structure of red rice germplasm in Bangladesh. PLoS One 13(5):e0196096
Jain S, Jain RK, McCOUCH SR (2004) Genetic analysis of Indian aromatic and quality rice (Oryza sativa L.) germplasm using panels of fluorescently-labeled microsatellite markers. Theoretical Appl Genet 109(5): 965–977
Jasim Aljumaili S, Rafii MY, Latif MA, Sakimin SZ, Arolu IW, Miah G (2018) Genetic diversity of aromatic rice germplasm revealed by SSR markers. Biomed Res Int 2018:7658032
Jin L, Lu Y, Xiao P, Sun M, Corke H, Bao J (2010) Genetic diversity and population structure of a diverse set of rice germplasm for association mapping. Theor Appl Genet 121(3):475–487
Liu K, Muse SV (2005) Power Marker: an integrated analysis environment for genetic marker analysis. Bioinform Appl Note 21(9):2128–2129
Mason AS (2015) SSR genotyping. In: Plant genotyping (pp 77–89). Humana Press, New York, NY
Mondini L, Noorani A, Pagnotta MA (2009) Assessing plant genetic diversity by molecular tools. Diversity 1(1):19–35
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucl Acids Res 8(19):4321–4325
Onaga G, Egdane J, Edema R, Ismail AM (2013) Morphological and genetic diversity analysis of rice accessions (Oryza sativa L.) differing in Iron toxicity tolerance. Journal of Crop Science and Biotechnology 16(1):85–95
Perrier X, Flori A, Bonnot F (2003) Data analysis methods. In: Hamon P, Seguin M, Perrier X, Glaszmann JC (eds) Genetic diversity of cultivated tropical plants. Enfield Science Publishers, Montpellier, pp 43–76
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breeding 2(3):225–238
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Rana MM, Islam MA, Imran S, Ruban S, Hassan L (2018) Genetic diversity analysis of NERICA lines and parents using SSR markers. Int J Plant Soil Sci 23(6):1–10
Rashid MM, Imran S, Islam MA and Hassan L (2018) Genetic diversity analysis of rice landraces (Oryza sativa L.) for salt tolerance using SSR markers in Bangladesh. Fundamental and Applied Agriculture, 3(2):460–466
Rohmawati I, Nursusilawati P and Abdullah S (2021) Genetic diversity of Some Indonesian Local Rice Varieties based on Simple Sequence Repeat (SSR) marker related to Aromatic Genes. In IOP Conference Series: Earth and Environmental Science (Vol. 715, No. 1, p. 012046)
Sajib AM, Hossain MdM, Mosnaz ATMJ, Hossain H, Islam MdM, Ali MdS, Prodhan SH (2012) SSR marker-based molecular characterization and genetic diversity analysis of aromatic landraces of rice (Oryza sativa L.). Journal of Biological Science and Biotechnology 1(2):107–116
Singh N, Choudhury DR, Tiwari G, Singh AK, Kumar S, Srinivasan K, Singh R (2016) Genetic diversity trend in Indian rice varieties: an analysis using SSR markers. BMC Genet 17:127
Singha T, Mahamud MA, Imran S, Paul NC, Hoque MN, Chakrobarty T, Al Galib MA, Hassan L (2021) Genetic diversity analysis of advanced rice lines for salt tolerance using SSR markers. Asian J Med Biol Res 7(2):214–221
Sreenivasulu N, Butardo VM Jr, Misra G, Cuevas RP, Anacleto R, Kavi Kishor PB (2015) Designing climate-resilient rice with ideal grain quality suited for high-temperature stress. J Exp Bot 66(7):1737–1748
Sun J, Yang L, Wang J, Liu H, Zheng H, Xie D, Zhang M, Feng M, Jia Y, Zhao H and Zou D (2018) Identification of a cold-tolerant locus in rice (Oryza sativa L.) using bulked segregant analysis with a next-generation sequencing strategy. Rice, 11(1): 1–12
Toro MA, Caballero A (2005) Characterization and conservation of genetic diversity in subdivided populations. Philosophical Trans R Soc b 360(1459):1367–1378
Upadhyay P, Neeraja CN, Kole C, Singh VK (2012) Population structure and genetic diversity in popular rice varieties of India as evidenced from SSR analysis. Biochem Genet 50(9):770–783
Upadhyay P, Singh VK, Neeraja CN (2011). Identification of genotype-specific alleles and molecular diversity assessment of popular rice (Oryza sativa L.) varieties of India. International Journal of Plant Breeding and Genetics 5(2):130–140
Yogi R, Kumar N, Kumar R and Jain RK (2020) Genetic diversity analysis among important rice (Oryza sativa L.) genotypes using SSR markers. Advances in Bioresearch 11(2):68–74
Yu SB, Xu WJ, Vijayakumar CHM, Ali J, Fu BY, Xu JL, Jiang YZ, Marghirang R, Domingo J, Aquino C and Virmani SS (2003) Molecular diversity and multilocus organization of the parental lines used in the International Rice Molecular Breeding Program. Theoretical and Applied Genetics, 108(1):131–140
Zhao W, Chung JW, Ma KH, Kim TS, Kim SM, Shin DI, Kim CH, Koo HM, Park YJ (2009) Analysis of genetic diversity and population structure of rice cultivars from Korea, China and Japan using SSR markers. Genes & Genomics 31(4):283–292
Author Information
Plant Genetic Resources and Improvement Division, CSIR, National Botanical Research Institute, Lucknow, India