Phenotypic variability, path analysis and molecular diversity analysis in chickpea (Cicer arietinum L.)
Research Articles | Published: 18 April, 2019
First Page: 167
Last Page: 180
Views: 3869
Keywords: Chickpea, Genetic advance, Heritability, SSR, Path analysis, Variability
Abstract
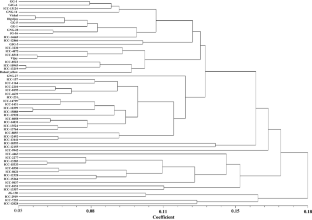
The existence and degree of genetic variability and its documentation in a gene-pool is obligatory in plant breeding. Along with variability, the comprehension of genetic parameters is indispensible for understanding and its administration during crop improvement. With this opinion, 12 phenotypic traits, one biochemical parameter and 23 microsatellite markers were used to examine the genetic variability in 58 chickpea genotypes. With immense heritability (> 60.20%), the genotypes exhibited lavish variability for most of the traits. Secondary branches per plant, pods per plant, seeds per pod, seeds per plant, hundred seed weight, seed yield per plant and harvest index demonstrated a high PCV than GCV. Seed yield per plant illustrated a significant genotypic level association with pods per plant (0.728**), seeds per plant (0.648**), 100 seed weight (0.338**) and harvest index (0.683**). Pods per plant (0.198), seeds per plant (0.672), harvest index (0.170) and 100 seed weight (0.665) showed significant direct effect on seed yield per plant during path analysis. Manhattan distance produced seven clusters, at cut-off value of 0.15, during phenotypic based clustering. Microsatellite markers amplified 296 loci. The polymorphic information content confined between 0.32 (TR3) − 0.93 (CAM0443) with an average of 0.83. The DNA marker based clustering generated three discrete clusters. Faint correlation (0.097) was found between the Manhattan’s and Nei’s distance. The outcomes of the current experiment advocated that both phenotypic as well as DNA markers should be practiced jointly to arrest the true genetic diversity and to reap heterosis during hybridization.
References
- Ali Q, Tahir MHN, Sadaqat HA, Arshad S, Farooq J, Ahsan M, Waseem M, Iqbal A (2011) Genetic variability and correlation analysis for quantitative traits in chickpea genotypes (Cicer arietinum L.). J Bact Res 3:6–9
- Anbessa Y, Warkentin T, Vandenberg A, Bandara M (2006) Heritability and predicted gain from selection in components of crop duration in divergent chickpea cross populations. Euphytica 152:1–8
- Archak S, Tyagia RK, Harerb PN, Mahaseb LB, Singh N, Dahiyaa OP, Nizarc MA, Singh M, Tilekar V, Kumar V, Dutta M, Singh NP, Bansala KC (2016) Characterization of chickpea germplasm conserved in the Indian National Gene bank and development of a core set using qualitative and quantitative trait data. Crop J 4:417–424
- Babbar A, Prakash V, Tiwari P, Iquebal MA (2012) Genetic variability in chickpea (Cicer arietinum L.) under late sown season. Leg Res 35(1):1–7
- Babbar A, Pandey S, Singh R (2015) Genetic studies on chickpea genotypes grown in late sown under rice fallow conditions of Madhya Pradesh. Electron J Pl Breed 6(3):738–748
- Boghara MC, Dhaduk HL, Kumar S, Parekh MJ, Patel NJ, Sharma R (2016) Genetic divergence, path analysis and molecular diversity analysis in cluster bean (Cyamopsis tetragonoloba L. Taub.). Int Crop Prod 89:468–477
- Burton GW (1952) Quantitative inheritance in grasses. In: Proceedings of the 6th international grassland congress, vol 1, pp 277–283
- Choudhary S, Kaur J, Chhuneja P, Sandhu JS, Singh I, Singh S, Sirari A (2013) Assessment of genetic diversity in kabuli chickpea (Cicer arietinum L.) genotypes in relation to seed size using SSR markers. J Food Leg 26(1 & 2):96–99
- Ciftci V, Togay N, Togay Y, Dogan Y (2004) Determining relationships among yield and some yield components using path coefficient analysis in chickpea. Asian J Pl Sci 3:632–635
- Dewey JR, Lu KH (1959) A correlation and path coefficient analysis of component of crested wheat seed production. Agron J 51:515–518
- Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
- Falconer DS (1960) Introduction to quantitative genetics. Oliver and Boyd, Edinburgh
- Farshadfar M, Farshadfar E (2008) Genetic variability and path analysis of chickpea (Cicer arietinum L.) landraces and line. J Appl Sci 8(21):3951–3956
- Ghaffari P, Talebi R, Keshavarzi F (2014) Genetic diversity and geographical differentiation of Iranian landrace, cultivars, and exotic chickpea lines as revealed by morphological and microsatellite markers. Physiol Mol Biol Plants 20(2):225–233
- Goldman D, Merril CR (1982) Silver staining of DNA in polyacrylamide gels: linearity and effect of fragment size. Electrophoresis 3:24–26
- Gowda CLL, Upadhyaya HD, Dronavalli N, Singh S (2011) Identification of large-seeded high-yielding stable kabuli chickpea germplasm lines for use in crop improvement. Crop Sci 51(1):198–209
- Hajibarat Z, Saidi A, Hajibarat Z, Talebi R (2014) Genetic diversity and population structure analysis of landrace and improved chickpea (Cicer arietinum L.) genotypes using morphological and microsatellite markers. Environ Exp Biol 12:161–166
- Iruela M, Rubio J, Cubero JI, Gil J, Millan T (2002) Phylogenetic analysis in the genus Cicer and cultivated chickpea using RAPD and ISSR markers. Theor Appl Genet 104(4):643–651
- Jafari N, Behroozi R, Bagheri A, Moshtaghi N (2012) Determination of genetic diversity of cultivated chickpea (Cicer arietinum L) using Medicago truncatula EST-SSRs. J Pl Mol Breed 1(2):1–16
- Jeena AS, Arora PP (2002) Path analysis in relation to selection in chickpea. Agril Sci Digest 22(2):122–127
- Jivani JV, Maheta DR, Vaddoria MA, Raval L (2013) Correlation and path coefficient analysis in chickpea (Cicer arietinum L.). Electron J Pl Breed 4(2):1167–1170
- Kalve S, Tadege M (2017) A comprehensive technique for artificial hybridization in Chickpea (Cicer arietinum). Plant Methods 13(52):1–9
- Kobraee S, Kayvan S, Behrooz R, Saeed K (2010) Investigation of correlation analysis and relationships between grain yield and other quantitative traits in chickpea. Afr J Biotech 9(6):2342–2348
- Kumar S, Ramavtar S, Vinod K, Govind V, Abhishek R (2013) Combining molecular-marker and chemical analysis of Capparis decidua (Capparaceae) in the Thar Desert of Western Rajasthan (India). Rev Biol Trop 61:311–320
- Liu K, Muse SV (2005) Power marker: integrated analysis environment for genetic markers data. Bioinfo 21:2128–2129
- Macar TK, Macar O, Mart Dİ (2017) Variability in some biochemical and nutritional characteristics in desi and Turkish kabuli chickpea (Cicer arietinum L.) types. Celal Bayar Üniversitesi Fen Bilimleri Dergisi 13(3):677–680
- Mahiboobsa M, Mannur DM, Shankergoud I, Somasekhar SG, Nidagundi JM (2017) Genetic variability studies in marker assisted backcross lines of chickpea (C. arietinum L). Int J Curr Microbiol Appl Sci 6(12):5375–5384
- Mehmet A, Emel GB (2013) Correlations and path analysis of yield and some yield components in chickpea (Cicer arietinum L.). J Food Agric Environ 11(3 & 4):748–749
- Mohammad RN, Sajad RM, Gomez H (2012) Genetic diversity in Iranian chickpea (Cicer arietinum L.) landraces as revealed by microsatellite markers. Czech J Genet Pl Breed 48(3):131–138
- Muhammad AA, Nausherwan NN, Amjad A, Zulkiffal M, Sajjad M (2009) Evaluation of selection criteria in Cicer arietinum L. using correlation coefficients and path analysis. Aust J Crop Sci 3(2):65–70
- Muhammad SS, Muhammad A, Zafar M, Muhammad A, Awais S, Muhammad IA (2016) Genetic variability and interrelationship of various agronomic traits using correlation and path analysis in chickpea (Cicer arietinum L.). Acad J Agril Res 4(2):82–85
- Naghavi MR, Jahansouz MR (2005) Variation in the agronomic and morphological traits of Iranian chickpea accessions. J Integr Plant Biol 47(3):375–379
- Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci 70(12):3321–3323
- Nguyen TT, Taylor PWJ, Redden RJ, Ford R (2004) Genetic diversity estimates in Cicer using AFLP analysis. Plant Breed 123:173–179
- Nkonge C, Ballance GM (1982) A sensitive colorimetric procedure for nitrogen determination in micro-Kjeldahl digests. J Agric Food Chem 30(3):416–420
- Noor F, Ashraf M, Gafoor A (2003) Path analysis and relationship among quantitative traits in Chickpea. Pak J Biol Sci 6(6):551–555
- Ozdemir S (1996) Path coefficient analysis for yield and its components in chickpea. ICPN 3:19–21
- Padmavathi PV, Murthy SS, Rao VS, Ahamed ML (2013) Correlation and path coefficient analysis in kabuli chickpea (Cicer arietinum L.). Int J Appl Biol Pharm 4(3):107–110
- Parameshwarappa SG, Palakshappa MG, Salimath PM, Parameshwarappa KG (2009) Evaluation and characterization of germplasm accessions of sesame (Sesamum indicum L.) Karnataka. J Agric Sci 22:1084–1086
- Parida G, Saghfi S, Eivazi A, Akbarzadeh A, Kavetskyy T, Aliyeva I, Khalilov R (2018) Study of genetic advance and broad sense heritability for grain yield and yield components of chickpea (Cicer arietinum L.) genotypes. Adv Biol Earth Sci 3(1):5–12
- Rathore PS, Sharma SK (2003) Scientific pulse production. Yash Publishing House, Bikaner, p 92
- Robinson HF, Comstock RE, Harvey PH (1949) Estimates of heritability and degree of dominance in corn. Agron J 41:353
- Roorkiwal M, Sawargaonkar SL, Chitikineni A, Thudi M, Saxena RK, Upadhyaya HD, Vales MI, Riera-Lizarazu O, Varshney RK (2013) Single nucleotide polymorphism genotyping for breeding and genetics applications in Chickpea and Pigeon pea using the BeadXpress Platform. Plant Genome 2(6):1–10
- Rukhsar Patel MP, Parmar DJ, Kalola AD, Kumar S (2017) Morphological and molecular diversity patterns in castor germplasm accessions. Ind Crops Prod 97:316–323
- Sachdeva S, Bharadwaj C, Sharma V, Patil BS, Soren KR, Roorkiwal M, Varshney R, Bhat KV (2018) Molecular and phenotypic diversity among chickpea (Cicer arietinum) genotypes as a function of drought tolerance. Crop Pasture Sci 2(69):142–153
- Saiyad MM, Kumar S (2017) Evaluation of maize genotypes for fodder quality traits and SSR diversity. J Plant Biochem Biotechnol 15:7. https://doi.org/10.1007/s13562-017-0418-6
- Sarvaliya VM (1993) Correlation, path analysis, selection indices and genetic divergence in chickpea. M.Sc. (Agri.) thesis submitted to Gujarat Agricultural University, Sardar Krushinagar
- Savaliya JJ, Kavani RH, Vaghela MD, Poshiya VK, Davada BK (2009) Genetic variability studies in kabuli chickpea (Cicer arietinum L.). Leg Res 32(3):191–194
- Sharanappa SD, Kumar J, Meena HP, Bharadwaj C, Jagadeesh HM, Raghvendra KP, Singode A (2014) Studies on heritability and genetic advance in Chickpea (Cicer arietinum L.). J Food Legumes 27(1):71–73
- Sharifi P, Astereki H, Pouresmael M (2018) Evaluation of variations in chickpea (Cicer arietinum L.) yield and yield components by multivariate technique. Ann Agrar Sci 16:136–142
- Singh T (2016) Estimation of genetic parameters and character associations for yield and quality traits in chickpea. Indian J Agric Res 50(2):112–117
- Singh SP, Sharma PC, Kumar R (2007) Correlation and path coefficient analysis in chickpea (Cicer arietinum L.). Int J Plant Sci 2(1):1–4
- Singh TP, Raiger HL, Kumari J, Singh A, Deshmukh PS (2014) Evaluation of chickpea genotypes for variability in seed protein content and yield components under restricted soil moisture condition. Indian J Plant Physiol 19(3):273–280
- Sudupak MA (2013) SSR-based genetic diversity assessment of Turkish Chickpea varieties. Biotechnol Biotechnol Equip 27(5):4087–4090
- Tadesse M, Asnake F, Million E, Nigusie G, Korbu L, Mohamed R, Dagnachew B, Assefa F, Chris OO (2016) Correlation and path coefficient analysis for various quantitative traits in desi chickpea genotypes under rainfed conditions in Ethiopia. J Agril Sci 8:112–118
- Talebi R, Fayaz F, Jelodar NA (2007) Correlation and path coefficient analysis of yield and yield components of chickpea (Cicer arietinum L.) under dry land condition in the west of Iran. Asian J Plant Sci 6(7):1151–1154
- Talekar SC, Viswanatha KP, Lohithaswa HC (2017) Assessment of genetic variability, character association and path analysis in F2 segregating population for quantitative traits in Chickpea. Int J Curr Microbiol Appl Sci 6(12):2184–2192
- Tesfamichae SM, Githiri SM, Nyende AB, Rao NVPRG (2015) Variation for agro-morphological traits among Kabuli chickpea (Cicer arietinum L) genotypes. J Agric Sci 7(7):75–92
- Thakur SK, Sirohi A (2009) Correlation and path coefficient Analysis in chickpea (Cicer arietinum L.) under different seasons. Leg Res 32(1):1–6
- Thakur NR, Toprope VN, Phanindra KS (2018) Estimation of genetic variability, correlation and path analysis for yield and yield contributing traits in chickpea (Cicer arietinum L). Leg Res 7(2):2298–2304
- Thudi M, Gaur PM, Krishnamurthy L, Mir RR, Kudapa H, Fikre A, Kimurto P, Tripathi S, Soren KR, Mulwa R, Bharadwaj C (2014) Genomics-assisted breeding for drought tolerance in chickpea. Funct Plant Biol 41(11):1178–1190
- Udupa SM, Robertson LD, Weigand F, Baum M, Kahl G (1999) Allelic variation at (TAA)n microsatellite loci in a world collection of chickpea (Cicer arietinum L.) germplasm. Mol General Genet 261(2):354–363
Author Information
Department of Genetics and Plant Breeding, B. A. College of Agriculture, Anand Agricultural University, Anand, India